5EAV
 
 | | Unliganded structure of the ornithine aminotransferase from Toxoplasma gondii | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unliganded structure of the ornithine aminotransferase from Toxoplasma gondii
To Be Published
|
|
5TFM
 
 | | Crystal Structure of Human Cadherin-23 EC6-8 | | Descriptor: | CALCIUM ION, Cadherin-23, SODIUM ION | | Authors: | Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2016-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5T2R
 
 | |
5TFK
 
 | |
5TFL
 
 | | Crystal Structure of Mouse Cadherin-23 EC7+8 | | Descriptor: | CALCIUM ION, Cadherin-23, SODIUM ION | | Authors: | Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2016-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
1KN2
 
 | | CATALYTIC ANTIBODY D2.3 COMPLEX | | Descriptor: | IG ANTIBODY D2.3 (HEAVY CHAIN), IG ANTIBODY D2.3 (LIGHT CHAIN), PARA-NITROPHENYL PHOSPHONOBUTANOYL L-ALANINE, ... | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Remarkable remote chiral recognition in a reaction mediated by a catalytic antibody.
J.Am.Chem.Soc., 124, 2002
|
|
1HEY
 
 | |
1ZTR
 
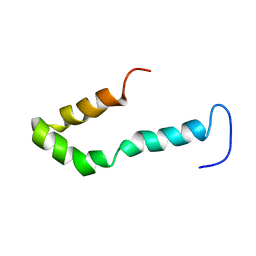 | | Solution structure of Engrailed homeodomain L16A mutant | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L, Markson, J.S, Mayor, U, Freund, S.M.V, Fersht, A.R. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a protein denatured state and folding intermediate.
Nature, 437, 2005
|
|
1QSL
 
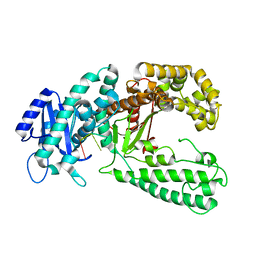 | |
1Z2B
 
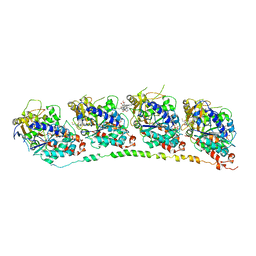 | | Tubulin-colchicine-vinblastine: stathmin-like domain complex | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-MERCAPTO-N-[1,2,3,10-TETRAMETHOXY-9-OXO-5,6,7,9-TETRAHYDRO-BENZO[A]HEPTALEN-7-YL]ACETAMIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gigant, B, Wang, C, Ravelli, R.B.G, Roussi, F, Steinmetz, M.O, Curmi, P.A, Sobel, A, Knossow, M. | | Deposit date: | 2005-03-08 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural basis for the regulation of tubulin by vinblastine.
Nature, 435, 2005
|
|
4HNA
 
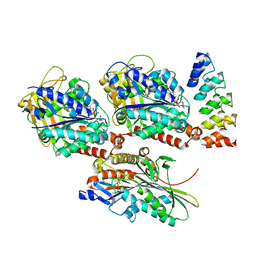 | |
1ZMD
 
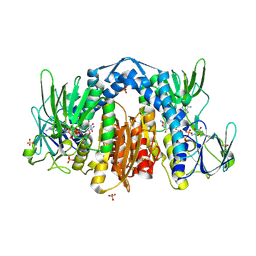 | | Crystal Structure of Human dihydrolipoamide dehydrogenase complexed to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of Human Dihydrolipoamide Dehydrogenase: NAD+/NADH Binding and the Structural Basis of Disease-causing Mutations
J.Mol.Biol., 350, 2005
|
|
2AXM
 
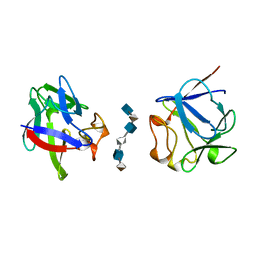 | | HEPARIN-LINKED BIOLOGICALLY-ACTIVE DIMER OF FIBROBLAST GROWTH FACTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Digabriele, A.D, Lax, I, Chen, D.I, Svahn, C.M, Jaye, M, Schlessinger, J, Hendrickson, W.A. | | Deposit date: | 1997-10-20 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a heparin-linked biologically active dimer of fibroblast growth factor.
Nature, 393, 1998
|
|
2FQY
 
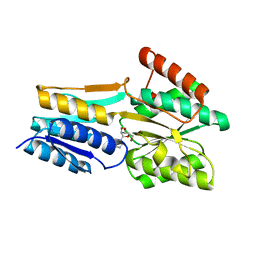 | | PnrA from Treponema pallidum complexed with adenosine. | | Descriptor: | ADENOSINE, Membrane lipoprotein tmpC | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Machius, M, Norgard, M.V. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum
J.Biol.Chem., 281, 2006
|
|
1ZMC
 
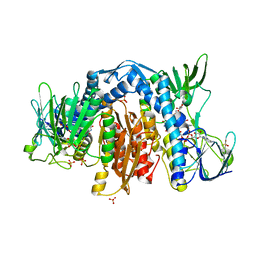 | | Crystal Structure of Human dihydrolipoamide dehydrogenase complexed to NAD+ | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal Structure of Human Dihydrolipoamide Dehydrogenase: NAD(+)/NADH Binding and the Structural Basis of Disease-causing Mutations
J.Mol.Biol., 350, 2005
|
|
3B3O
 
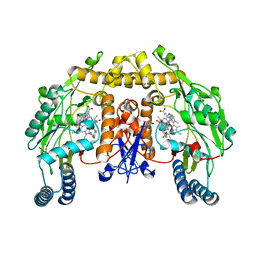 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-AMINO-4-METHYLPYRIDIN-2-YL)METHYL]PYRROLIDIN-3-YL}-N'-(4-CHLOROBENZYL)ETHANE-1,2-DIAMINE, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
4L71
 
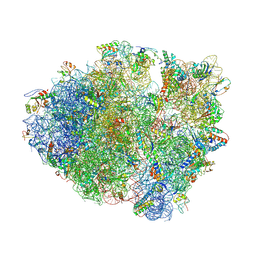 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-A on the Ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-13 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.900001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4L47
 
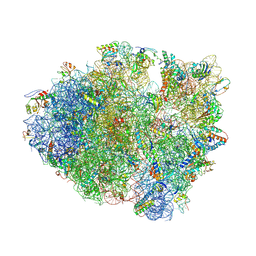 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.220001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LFZ
 
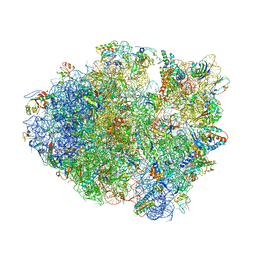 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U in the Absence of Paromomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-27 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.92000055 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2F60
 
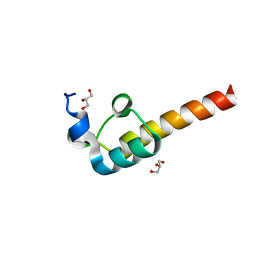 | | Crystal Structure of the Dihydrolipoamide Dehydrogenase (E3)-Binding Domain of Human E3-Binding Protein | | Descriptor: | GLYCEROL, Pyruvate dehydrogenase protein X component | | Authors: | Brautigam, C.A, Chuang, J.L, Wynn, R.M, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-11-28 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insight into Interactions between Dihydrolipoamide Dehydrogenase (E3) and E3 Binding Protein of Human Pyruvate Dehydrogenase Complex.
Structure, 14, 2006
|
|
1PK8
 
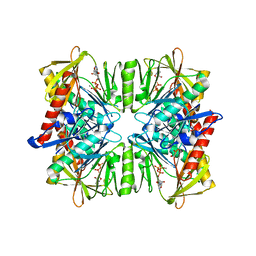 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-06-05 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
2EYY
 
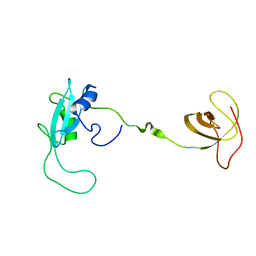 | | CT10-Regulated Kinase isoform I | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1I56
 
 | |
2EYV
 
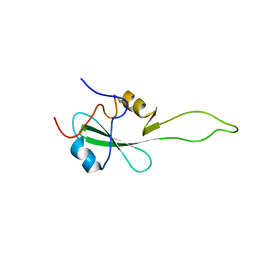 | | SH2 domain of CT10-Regulated Kinase | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2GSY
 
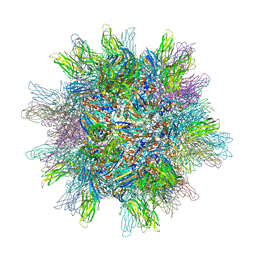 | | The 2.6A structure of Infectious Bursal Virus Derived T=1 Particles | | Descriptor: | CALCIUM ION, polyprotein | | Authors: | Garriga, D, Querol-Audi, J, Abaitua, F, Saugar, I, Pous, J, Verdaguer, N, Caston, J.R, Rodriguez, J.F. | | Deposit date: | 2006-04-27 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6-angstrom structure of infectious bursal disease virus-derived t=1 particles reveals new stabilizing elements of the virus capsid.
J.Virol., 80, 2006
|
|
