4U6K
 
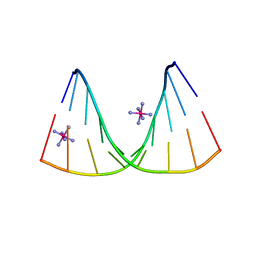 | | Crystal structure of DNA/RNA duplex containing 2'-4'-BNA-NC | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*(NCU)P*(NTT)P*CP*TP*TP*CP*TP*(NTT)P*(NCU))-3'), RNA (5'-R(*GP*AP*AP*GP*AP*AP*GP*AP*G)-3') | | Authors: | Kondo, J, Nomura, Y, Kitahara, Y, Obika, S, Torigoe, H. | | Deposit date: | 2014-07-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a 2',4'-BNA(NC)[N-Me]-modified antisense gapmer in complex with the target RNA.
Chem.Commun.(Camb.), 52, 2016
|
|
4U6M
 
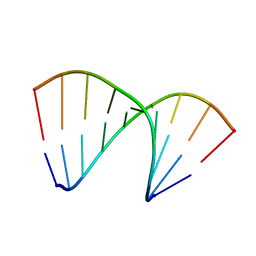 | | Crystal structure of DNA/RNA duplex obtained in the presence of Spermine | | Descriptor: | DNA (5'-D(*(5CM)P*TP*CP*TP*TP*CP*TP*TP*(5CM))-3'), RNA (5'-R(*GP*AP*AP*GP*AP*AP*GP*AP*G)-3') | | Authors: | Kondo, J, Nomura, Y, Kitahara, Y, Obika, S, Torigoe, H. | | Deposit date: | 2014-07-29 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a 2',4'-BNA(NC)[N-Me]-modified antisense gapmer in complex with the target RNA.
Chem.Commun.(Camb.), 52, 2016
|
|
4U8A
 
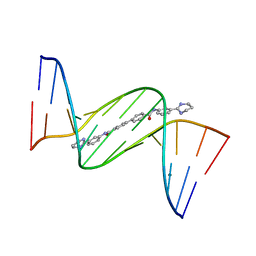 | | Crystal structure of D(CGCGAATTCGCG)2 complexed with BPH-1503 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, N,N'-bis[3-(1,4,5,6-tetrahydropyrimidin-2-yl)phenyl]biphenyl-4,4'-dicarboxamide | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Antibacterial drug leads: DNA and enzyme multitargeting.
J.Med.Chem., 58, 2015
|
|
4U8D
 
 | |
4UAP
 
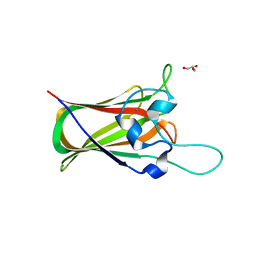 | | X-ray structure of GH31 CBM32-2 bound to GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Grondin, J.M, Abe, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-08-11 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
4U1Z
 
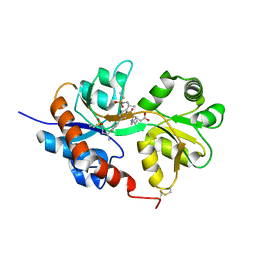 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form D | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9401 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U22
 
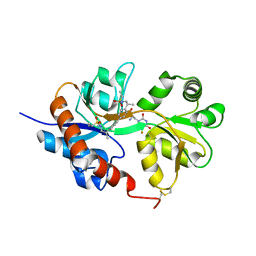 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form D | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4409 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U23
 
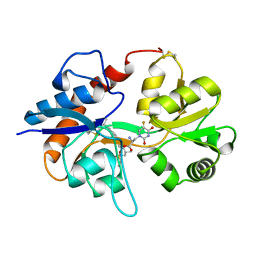 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form F | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6734 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4UAM
 
 | | 1.8 Angstrom crystal structure of IMP-1 metallo-beta-lactamase with a mixed iron-zinc center in the active site | | Descriptor: | CITRATE ANION, FE (III) ION, IMP-1 metallo-beta-lactamase, ... | | Authors: | Carruthers, T.J, Carr, P.D, Jackson, C.J, Otting, G. | | Deposit date: | 2014-08-11 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Iron(III) Located in the Dinuclear Metallo-beta-Lactamase IMP-1 by Pseudocontact Shifts.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4U2M
 
 | |
4U2Y
 
 | | Sco GlgEI-V279S in Complex with Reaction Intermediate Azasugar | | Descriptor: | (2R,3R,4R,5R)-4-hydroxy-2,5-bis(hydroxymethyl)pyrrolidin-3-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1 | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4UA4
 
 | |
4UBH
 
 | | Resting state of rat cysteine dioxygenase Y157F variant | | Descriptor: | CHLORIDE ION, Cysteine dioxygenase type 1, FE (II) ION, ... | | Authors: | Tchesnokov, E.P, Fellner, M, Jameson, G.N.L, Wilbanks, S.M. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of cysteine dioxygenase mutant
To be published
|
|
4U40
 
 | |
4U42
 
 | | MAP4K4 T181E Mutant Bound to inhibitor compound 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(3S)-3-(4-methyl-1H-pyrazol-3-yl)piperidin-1-yl]pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U2N
 
 | |
4U3Z
 
 | | APO MAP4K4 T181E Phosphomimetic Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase kinase kinase kinase 4, SODIUM ION | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U4E
 
 | |
4U4H
 
 | |
4U4J
 
 | |
4U5L
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzyl]-L-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U5V
 
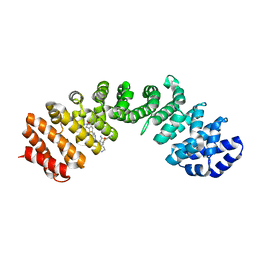 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-{[5-(pyridin-3-yl)thiophen-2-yl]methyl}-L-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U6O
 
 | |
4U7G
 
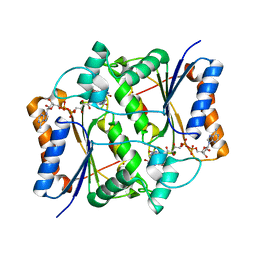 | | Oxidized quinone reductase 2 in complex with CK2 inhibitor TBBz | | Descriptor: | 4,5,6,7-TETRABROMO-BENZIMIDAZOLE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Quinone Reductase 2 Is an Adventitious Target of Protein Kinase CK2 Inhibitors TBBz (TBI) and DMAT.
Biochemistry, 54, 2015
|
|
4U7F
 
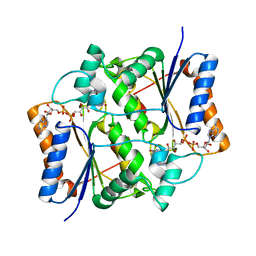 | | Reduced quinone reductase 2 in complex with CK2 inhibitor DMAT | | Descriptor: | 4,5,6,7-TETRABROMO-N,N-DIMETHYL-1H-BENZIMIDAZOL-2-AMINE, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Leung, K.K, Shilton, B.H. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Quinone Reductase 2 Is an Adventitious Target of Protein Kinase CK2 Inhibitors TBBz (TBI) and DMAT.
Biochemistry, 54, 2015
|
|
