3LW5
 
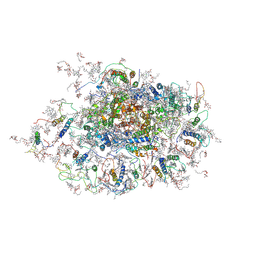 | | Improved model of plant photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3g54890, BETA-CAROTENE, ... | | Authors: | Nelson, N, Toporik, H. | | Deposit date: | 2010-02-23 | | Release date: | 2010-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure determination and improved model of plant photosystem I
J.Biol.Chem., 285, 2010
|
|
3LWB
 
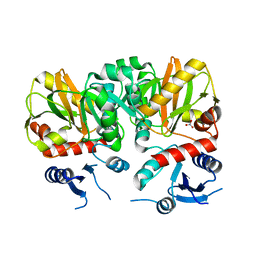 | | Crystal Structure of apo D-alanine:D-alanine Ligase (Ddl) from Mycobacterium tuberculosis | | Descriptor: | D-alanine--D-alanine ligase, NITRATE ION | | Authors: | Bruning, J.B, Murillo, A.C, Chacon, O, Barletta, R.G, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Mycobacterium tuberculosis D-Alanine:D-Alanine Ligase, a Target of the Antituberculosis Drug D-Cycloserine.
Antimicrob.Agents Chemother., 55, 2011
|
|
3LYP
 
 | |
3M3X
 
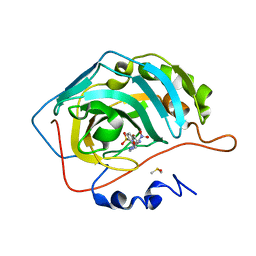 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{2-[N-(6-methoxy-5-nitropyrimidin-4-yl)amino]ethyl}benzenesulfonamide | | Descriptor: | 4-{2-[(6-methoxy-5-nitropyrimidin-4-yl)amino]ethyl}benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
3M0P
 
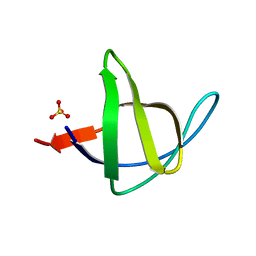 | |
3M0S
 
 | |
3M5E
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-{[N-(6-chloro-5-formyl-2-methylthiopyrimidin-4-yl)amino]methyl}benzenesulfonamide | | Descriptor: | 4-({[6-chloro-5-formyl-2-(methylsulfanyl)pyrimidin-4-yl]amino}methyl)benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-[N-(Substituted 4-pyrimidinyl)amino]benzenesulfonamides as inhibitors of carbonic anhydrase isozymes I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
3M9F
 
 | | HIV protease complexed with compound 10b | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-[(1S,5S)-5-{[(4-aminophenyl)sulfonyl](3-methylbutyl)amino}-1-methyl-6-oxohexyl]-Nalpha-(methoxycarbonyl)-beta-phenyl-L-phenylalaninamide | | Authors: | Su, H.P. | | Deposit date: | 2010-03-22 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Epsilon substituted lysinol derivatives as HIV-1 protease inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3M82
 
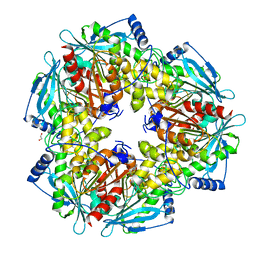 | |
3MC3
 
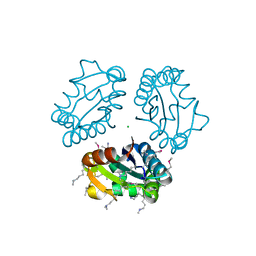 | |
3M98
 
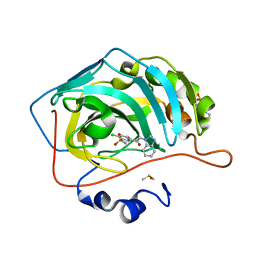 | |
3MCQ
 
 | |
3JXA
 
 | | Immunoglobulin domains 1-4 of mouse CNTN4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin 4 | | Authors: | Bouyain, S. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JYH
 
 | | Human dipeptidyl peptidase DPP7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl-peptidase 2, ... | | Authors: | Dobrovetsky, E, Dong, A, Seitova, A, Crombett, L, Paganon, S, Cossar, D, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Hassel, A, Shewchuk, L, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Human dipeptidyl peptidase DPP7
To be Published
|
|
3JZT
 
 | | Structure of a cubic crystal form of X (ADRP) domain from FCoV with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wojdyla, J.A, Manolaridis, I, Tucker, P.A. | | Deposit date: | 2009-09-24 | | Release date: | 2010-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Structure of the X (ADRP) domain of nsp3 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3K0I
 
 | | Crystal structure of Cu(I)CusA | | Descriptor: | COPPER (I) ION, Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.116 Å) | | Cite: | Crystal structure of CusA
To be Published
|
|
3K6W
 
 | | Apo and ligand bound structures of ModA from the archaeon Methanosarcina acetivorans | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3K75
 
 | |
3K7O
 
 | |
3K7S
 
 | |
3K8N
 
 | |
3K8Q
 
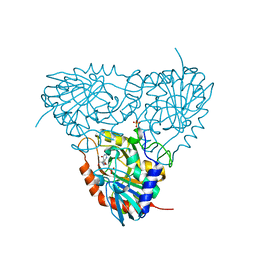 | | Crystal structure of human purine nucleoside phosphorylase in complex with SerMe-Immucillin H | | Descriptor: | 7-({[2-hydroxy-1-(hydroxymethyl)ethyl]amino}methyl)-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ho, M, Almo, S.C, Scharmm, V.L. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human nucleoside phosphorylase in complex with SerMe-ImmH
Proc.Natl.Acad.Sci.USA, 2010
|
|
3K8Z
 
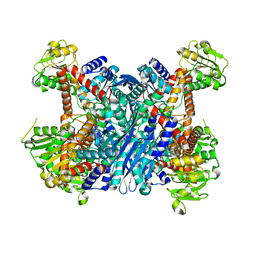 | | Crystal Structure of Gudb1 a decryptified secondary glutamate dehydrogenase from B. subtilis | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Gunka, K, Newman, J.A, Commichau, F.M, Herzberg, C, Rodrigues, C, Hewitt, L, Lewis, R.J, Stulke, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional dissection of a trigger enzyme: mutations of the bacillus subtilis glutamate dehydrogenase RocG that affect differentially its catalytic activity and regulatory properties
J.Mol.Biol., 400, 2010
|
|
3K9M
 
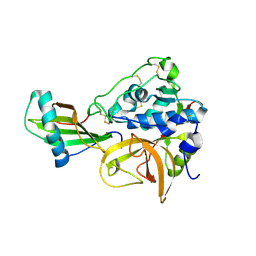 | | Cathepsin B in complex with stefin A | | Descriptor: | Cathepsin B, Cystatin-A | | Authors: | Renko, M, Turk, D. | | Deposit date: | 2009-10-16 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Stefin A displaces the occluding loop of cathepsin B only by as much as required to bind to the active site cleft
Febs J., 277, 2010
|
|
3JSP
 
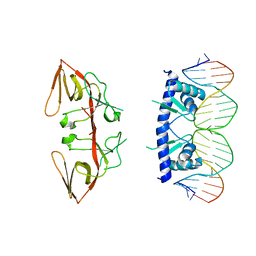 | |
