6EU3
 
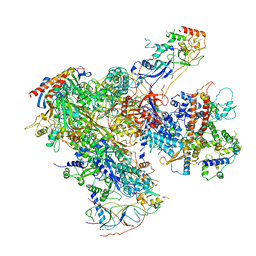 | | Apo RNA Polymerase III - closed conformation (cPOL3) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
5XON
 
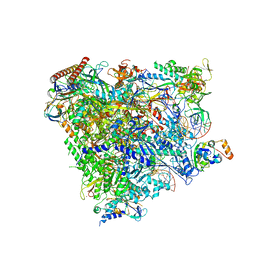 | | RNA Polymerase II elongation complex bound with Spt4/5 and TFIIS | | Descriptor: | DNA (48-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Yokoyama, T, Shigematsu, H, Shirouzu, M, Sekine, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of the complete elongation complex of RNA polymerase II with basal factors
Science, 357, 2017
|
|
6EXV
 
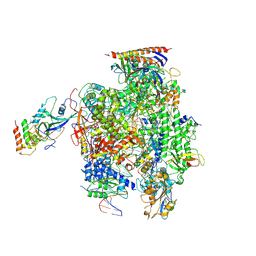 | | Structure of mammalian RNA polymerase II elongation complex inhibited by Alpha-amanitin | | Descriptor: | AMATOXIN, DNA (25-MER), DNA (36-MER), ... | | Authors: | Liu, X, Farnung, L, Wigge, C, Cramer, P. | | Deposit date: | 2017-11-09 | | Release date: | 2018-03-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of a mammalian RNA polymerase II elongation complex inhibited by alpha-amanitin.
J. Biol. Chem., 293, 2018
|
|
6YYS
 
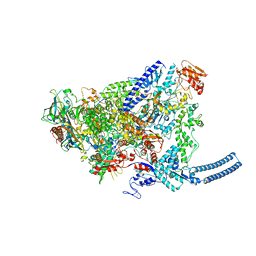 | | Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State II, primary channel engaged and active site interfering | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Koval, T, Krasny, L, Dohnalek, J. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mycobacterial HelD is a nucleic acids-clearing factor for RNA polymerase.
Nat Commun, 11, 2020
|
|
1TWG
 
 | | RNA polymerase II complexed with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
1TWH
 
 | | RNA polymerase II complexed with 2'dATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
1TWF
 
 | | RNA polymerase II complexed with UTP at 2.3 A resolution | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
1TWC
 
 | | RNA polymerase II complexed with GTP | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
1TWA
 
 | | RNA polymerase II complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
3AOI
 
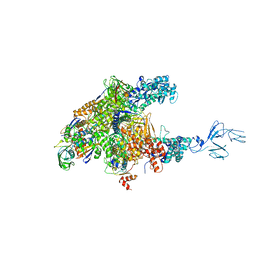 | | RNA polymerase-Gfh1 complex (Crystal type 2) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*A*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
3AOH
 
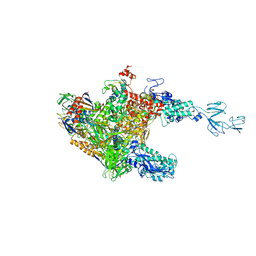 | | RNA polymerase-Gfh1 complex (Crystal type 1) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*AP*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-28 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
7OBB
 
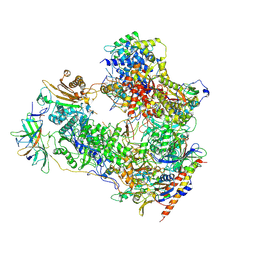 | | Cryo-EM structure of human RNA Polymerase I Open Complex | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OB9
 
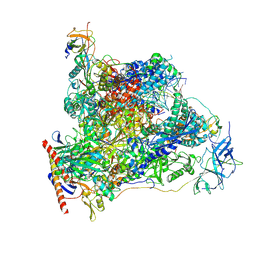 | | Cryo-EM structure of human RNA Polymerase I in elongation state | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OBA
 
 | | Cryo-EM structure of human RNA Polymerase I in complex with RRN3 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA1, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA2, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3N97
 
 | | RNA polymerase alpha C-terminal domain (E. coli) and sigma region 4 (T. aq. mutant) bound to (UP,-35 element) DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*CP*AP*TP*GP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*TP*TP*TP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*AP*AP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Lara-Gonzalez, S, Birktoft, J.J, Lawson, C.L. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.252 Å) | | Cite: | The RNA Polymerase alpha Subunit Recognizes the DNA Shape of the Upstream Promoter Element.
Biochemistry, 59, 2020
|
|
9BW0
 
 | | RNA Polymerase II - No ATP | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Calero, G. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6A5L
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5R
 
 | | RNA polymerase II elongation complex stalled at SHL(-2) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5T
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5P
 
 | | RNA polymerase II elongation complex stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5U
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA, tilt conformation | | Descriptor: | DNA (198-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5O
 
 | | RNA polymerase II elongation complex stalled at SHL(-6) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
5FLM
 
 | | Structure of transcribing mammalian RNA polymerase II | | Descriptor: | DNA, DNA-RNA ELONGATION SCAFFOLD, DNA-DIRECTED RNA POLYMERASE, ... | | Authors: | Bernecky, C, Herzog, F, Baumeister, W, Plitzko, J.M, Cramer, P. | | Deposit date: | 2015-10-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of Transcribing Mammalian RNA Polymerase II
Nature, 529, 2016
|
|
6INQ
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA (+1 position) | | Descriptor: | DNA (198-MER), DNA (31-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-10-26 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
8HE5
 
 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.95 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
