4PY7
 
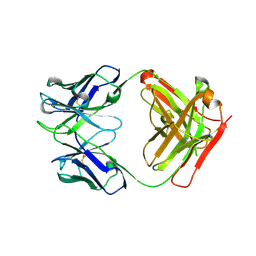 | | Crystal Structure of Fab 3.1 | | Descriptor: | antibody 3.1 heavy chain, antibody 3.1 light chain | | Authors: | Dreyfus, C. | | Deposit date: | 2014-03-26 | | Release date: | 2014-05-21 | | Last modified: | 2014-06-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alternative Recognition of the Conserved Stem Epitope in Influenza A Virus Hemagglutinin by a VH3-30-Encoded Heterosubtypic Antibody.
J.Virol., 88, 2014
|
|
3ODZ
 
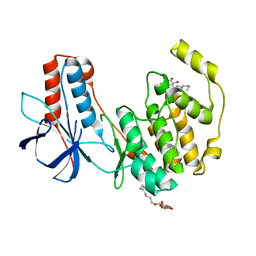 | | Crystal structure of P38alpha Y323R active mutant | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Livnah, O, Tzarum, N. | | Deposit date: | 2010-08-12 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active mutants of the TCR-mediated p38alpha alternative activation site show changes in the phosphorylation lip and DEF site formation.
J.Mol.Biol., 405, 2011
|
|
3OV9
 
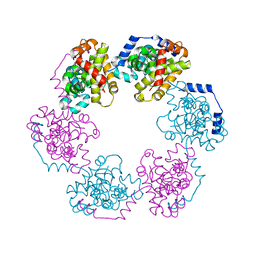 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein, SODIUM ION | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
2MTG
 
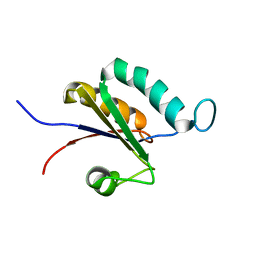 | | Solution structure of the RRM1 of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
7OA4
 
 | | Crystal structure of the N-terminal endonuclease domain of La Crosse virus L-protein bound to compound L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Feracci, M, Hernandez, S, Vincentelli, R, Ferron, F, Reguera, J, Canard, B, Alvarez, K. | | Deposit date: | 2021-04-19 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the N-terminal endonuclease domain of La Crosse virus L-protein bound to compound L-742,001
To Be Published
|
|
3K5M
 
 | | Crystal structure of E.coli Pol II-abasic DNA-ddGTP Lt(-2, 2) ternary complex | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*TP*CP*CP*TP*GP*(3DR)P*AP*CP*GP*CP*TP*AP*GP*GP*CP*AP*CP*A)-3'), ... | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II.
Cell(Cambridge,Mass.), 139, 2009
|
|
6IB7
 
 | | Structure of wild type SuhB | | Descriptor: | GLYCEROL, Inositol-1-monophosphatase, MAGNESIUM ION | | Authors: | Huang, Y.H, Loll, B, Wahl, M.C. | | Deposit date: | 2018-11-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis.
Nucleic Acids Res., 47, 2019
|
|
8A22
 
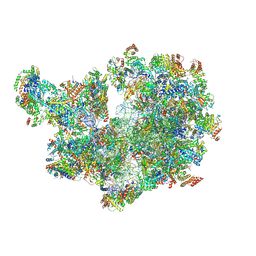 | | Structure of the mitochondrial ribosome from Polytomella magna | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Tobiasson, V, Berzina, I, Amunts, A. | | Deposit date: | 2022-06-02 | | Release date: | 2022-11-16 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structure of a mitochondrial ribosome with fragmented rRNA in complex with membrane-targeting elements.
Nat Commun, 13, 2022
|
|
4CYJ
 
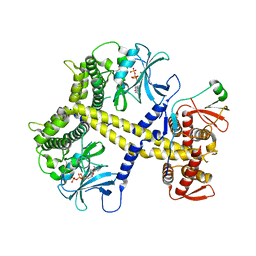 | | Chaetomium thermophilum Pan2:Pan3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-11 | | Last modified: | 2014-07-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
2KVI
 
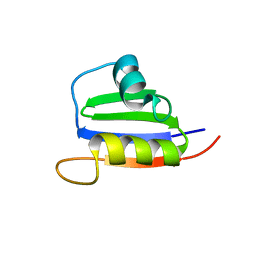 | | Structure of Nab3 RRM | | Descriptor: | Nuclear polyadenylated RNA-binding protein 3 | | Authors: | Pergoli, R, Kubicek, K, Hobor, F, Pasulka, J, Stefl, R. | | Deposit date: | 2010-03-15 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and RNA-binding study of RNA-recognition motif of Nab3
J.Biol.Chem., 2010
|
|
4J3K
 
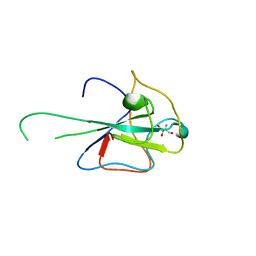 | |
1TRJ
 
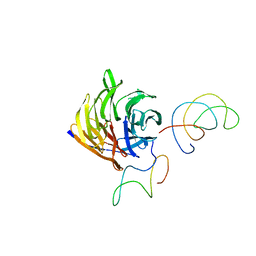 | | Homology Model of Yeast RACK1 Protein fitted into 11.7A cryo-EM map of Yeast 80S Ribosome | | Descriptor: | Guanine nucleotide-binding protein beta subunit-like protein, Helix 39 of 18S rRNA, Helix 40 of 18S rRNA | | Authors: | Sengupta, J, Nilsson, J, Gursky, R, Spahn, C.M, Nissen, P, Frank, J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM
Nat.Struct.Mol.Biol., 11, 2004
|
|
8QLP
 
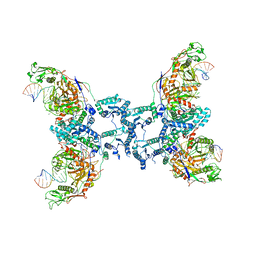 | | CryoEM structure of the RNA/DNA bound SPARTA (BabAgo/TIR-APAZ) tetrameric complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*C)-3'), MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
7K06
 
 | |
7K07
 
 | |
7K05
 
 | |
6P2H
 
 | |
2M1A
 
 | | HIV-1 Rev ARM peptide (residues T34-R50) | | Descriptor: | HIV-1 Rev arginine-rich motif (ARM) | | Authors: | Casu, F, Duggan, B.M, Hennig, M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Arginine-Rich RNA-Binding Motif of HIV-1 Rev Is Intrinsically Disordered and Folds upon RRE Binding.
Biophys.J., 105, 2013
|
|
3TD1
 
 | |
7NYQ
 
 | | Crystal structure of the Mei-P26 NHL domain | | Descriptor: | Meiotic P26, isoform C | | Authors: | Salerno-Kochan, A, Gaik, M, Medenbach, J, Glatt, S. | | Deposit date: | 2021-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into RNA recognition and gene regulation by the TRIM-NHL protein Mei-P26.
Life Sci Alliance, 5, 2022
|
|
4N0O
 
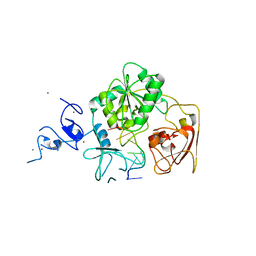 | | Complex structure of Arterivirus nonstructural protein 10 (helicase) with DNA | | Descriptor: | CALCIUM ION, DNA, Replicase polyprotein 1ab, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
7LPY
 
 | |
7LPZ
 
 | |
7LQ0
 
 | |
4A76
 
 | | The Lin28b Cold shock domain in complex with heptathymidine | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP)-3', LIN28 COLD SHOCK DOMAIN | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2011-11-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
