1L8L
 
 | | Molecular basis for the local confomational rearrangement of human phosphoserine phosphatase | | Descriptor: | D-2-AMINO-3-PHOSPHONO-PROPIONIC ACID, L-3-phosphoserine phosphatase | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase.
J.Biol.Chem., 277, 2002
|
|
1LKP
 
 | | Crystal structure of Desulfovibrio vulgaris rubrerythrin all-iron(II) form, azide adduct | | Descriptor: | AZIDE ION, FE (II) ION, Rubrerythrin | | Authors: | Jin, S, Kurtz Jr, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2002-04-25 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray Crystal Structures of Reduced Rubrerythrin and its Azide Adduct: A Structure-Based Mechanism for a Non-Heme Diiron Peroxidase
J.Am.Chem.Soc., 124, 2002
|
|
1MXD
 
 | | Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
1MXG
 
 | | Crystal Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei in complex with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
1N1D
 
 | | Glycerol-3-phosphate cytidylyltransferase complexed with CDP-glycerol | | Descriptor: | SULFATE ION, [CYTIDINE-5'-PHOSPHATE] GLYCERYLPHOSPHORIC ACID ESTER, glycerol-3-phosphate cytidylyltransferase | | Authors: | Pattridge, K.A, Weber, C.H, Friesen, J.A, Sankar, S, Kent, C, Ludwig, M.L. | | Deposit date: | 2002-10-17 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Glycerol-3-phosphate cytidylyltransferase. Structural changes induced by binding of CDP-glycerol and the role of lysine residues in catalysis
J.Biol.Chem., 278, 2003
|
|
1MSA
 
 | |
8B9X
 
 | | Chimeric protein of human UFM1 E3 ligase, UFL1, and DDRGK1 | | Descriptor: | CHLORIDE ION, DDRGK domain-containing protein 1,E3 UFM1-protein ligase 1 | | Authors: | Wiener, R, Isupov, M, banerjee, S. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.066 Å) | | Cite: | Structural study of UFL1-UFC1 interaction uncovers the role of UFL1 N-terminal helix in ufmylation.
Embo Rep., 24, 2023
|
|
5K39
 
 | | THE TYPE II COHESIN DOCKERIN COMPLEX FROM CLOSTRIDIUM THERMOCELLUM | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin module from a protein of unknown function | | Authors: | Viegas, A, Pinheiro, B, Bras, J.L.A, Romao, M.J, Alves, V, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
1SPA
 
 | | ROLE OF ASP222 IN THE CATALYTIC MECHANISM OF ESCHERICHIA COLI ASPARTATE AMINOTRANSFERASE: THE AMINO ACID RESIDUE WHICH ENHANCES THE FUNCTION OF THE ENZYME-BOUND COENZYME PYRIDOXAL 5'-PHOSPHATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, N-METHYL-4-DEOXY-4-AMINO-PYRIDOXAL-5-PHOSPHATE | | Authors: | Hinoue, Y, Yano, T, Metzler, D.E, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1993-01-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Asp222 in the catalytic mechanism of Escherichia coli aspartate aminotransferase: the amino acid residue which enhances the function of the enzyme-bound coenzyme pyridoxal 5'-phosphate.
Biochemistry, 31, 1992
|
|
1DJ1
 
 | | CRYSTAL STRUCTURE OF R48A MUTANT OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Goodin, D.B. | | Deposit date: | 1999-11-30 | | Release date: | 1999-12-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Unusual oxidative chemistry of N(omega)-hydroxyarginine and N-hydroxyguanidine catalyzed at an engineered cavity in a heme peroxidase.
J.Biol.Chem., 275, 2000
|
|
1ENC
 
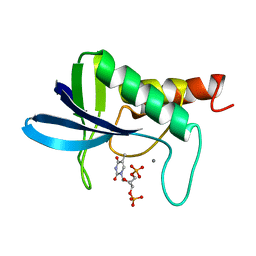 | | CRYSTAL STRUCTURES OF THE BINARY CA2+ AND PDTP COMPLEXES AND THE TERNARY COMPLEX OF THE ASP 21->GLU MUTANT OF STAPHYLOCOCCAL NUCLEASE. IMPLICATIONS FOR CATALYSIS AND LIGAND BINDING | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Libson, A, Gittis, A, Lattman, E.E. | | Deposit date: | 1994-02-14 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the binary Ca2+ and pdTp complexes and the ternary complex of the Asp21-->Glu mutant of staphylococcal nuclease. Implications for catalysis and ligand binding.
Biochemistry, 33, 1994
|
|
3S6M
 
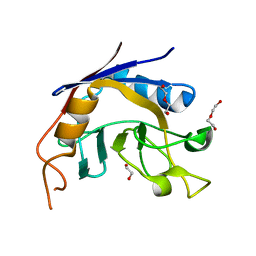 | |
1R1C
 
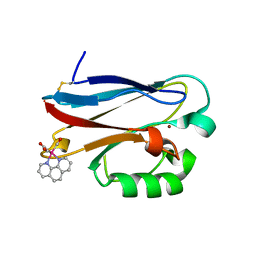 | | PSEUDOMONAS AERUGINOSA W48F/Y72F/H83Q/Y108W-AZURIN RE(PHEN)(CO)3(HIS107) | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (I) ION | | Authors: | Miller, J.E, Gradinaru, C, Crane, B.R, Di Bilio, A.J. | | Deposit date: | 2003-09-23 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spectroscopy and reactivity of a photogenerated tryptophan radical in a structurally defined protein environment
J.Am.Chem.Soc., 125, 2003
|
|
1EVW
 
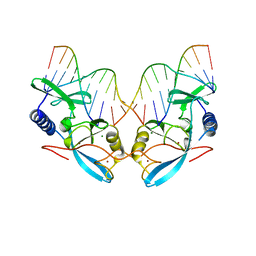 | | L116A MUTANT OF THE HOMING ENDONUCLEASE I-PPOI COMPLEXED TO HOMING SITE DNA. | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*TP*AP*CP*CP*TP*TP*AP*A)-3'), DNA (5'-D(P*GP*AP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
1O8F
 
 | |
8CJB
 
 | | A268M variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | ACETATE ION, CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coupling CO 2 Reduction and Acetyl-CoA Formation: The Role of a CO Capturing Tunnel in Enzymatic Catalysis.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7NA6
 
 | | Cryo-EM structure of AAV True Type | | Descriptor: | Capsid protein VP1 | | Authors: | Bennett, A.D, McKenna, R. | | Deposit date: | 2021-06-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Comparative structural, biophysical, and receptor binding study of true type and wild type AAV2.
J.Struct.Biol., 213, 2021
|
|
1RCJ
 
 | | Crystal structure of E166A mutant of SHV-1 beta-lactamase with the trans-enamine intermediate of tazobactam | | Descriptor: | Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, TAZOBACTAM TRANS-ENAMINE INTERMEDIATE | | Authors: | Padayatti, P.S, Helfand, M.S, Totir, M.A, Carey, M.P, Hujer, A.M, Carey, P.R, Bonomo, R.A, van den Akker, F. | | Deposit date: | 2003-11-04 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Tazobactam Forms a Stoichiometric trans-Enamine Intermediate in the E166A Variant of SHV-1 beta-Lactamase: 1.63 A Crystal Structure
Biochemistry, 43, 2004
|
|
8CGX
 
 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using XDS | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
3SNZ
 
 | |
8CGY
 
 | | Trypanosoma brucei IMP dehydrogenase (ori) crystallized in High Five cells reveals native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo; XDS processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD5
 
 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
5KHA
 
 | |
1NNA
 
 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | Descriptor: | CALCIUM ION, NEURAMINIDASE | | Authors: | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | Deposit date: | 1993-03-08 | | Release date: | 1994-04-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
6IPU
 
 | |
