2QJS
 
 | |
2QK2
 
 | |
3EJP
 
 | | Golgi alpha-Mannosidase II in complex with 5-substituted swainsonine analog: (5R)-5-[2'-oxo-2'-(phenyl)ethyl]-swainsonine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-phenyl-2-[(1S,2R,5R,8R,8aR)-1,2,8-trihydroxyoctahydroindolizin-5-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
3EEJ
 
 | |
2QKH
 
 | | Crystal structure of the extracellular domain of human GIP receptor in complex with the hormone GIP | | Descriptor: | Cyclic 2,3-di-O-methyl-alpha-D-glucopyranose-(1-4)-2-O-methyl-alpha-D-glucopyranose-(1-4)-2,6-di-O-methyl-alpha-D-glucopyranose-(1-4)-2-O-methyl-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-3-O-methyl-alpha-D-glucopyranose, D(-)-TARTARIC ACID, Glucose-dependent insulinotropic polypeptide, ... | | Authors: | Parthier, C, Kleinschmidt, M, Neumann, P, Rudolph, R, Manhart, S, Schlenzig, D, Fanghanel, J, Rahfeld, J.-U, Demuth, H.-U, Stubbs, M.T. | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3EF0
 
 | |
3TYT
 
 | |
2QLQ
 
 | | Crystal structure of SRC kinase domain with covalent inhibitor RL3 | | Descriptor: | (2E)-N-{4-[(3-bromophenyl)amino]quinazolin-6-yl}-4-(dimethylamino)but-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Michalczyk, A, Rode, H.B, Gruetter, C, Rauh, D. | | Deposit date: | 2007-07-13 | | Release date: | 2008-03-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insights into how irreversible inhibitors can overcome drug resistance in EGFR.
Bioorg.Med.Chem., 16, 2008
|
|
3TZN
 
 | |
3TPR
 
 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U1P
 
 | | Crystal Structure of M. tuberculosis LD-transpeptidase type 2 with Modified Catalytic Cysteine (C354) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Mycobacteria Tuberculosis LD-transpeptidase type 2 | | Authors: | Erdemli, S, Bianchet, M.A, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-09-30 | | Release date: | 2012-12-05 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Cell Wall of Mycobacterium tuberculosis: Structure and Mechanism of L,D-Transpeptidase 2.
Structure, 20, 2012
|
|
2QSZ
 
 | | Human nicotinamide riboside kinase 1 in complex with nicotinamide mononucleotide | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, CHLORIDE ION, Nicotinamide riboside kinase 1, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
2QOJ
 
 | | Coevolution of a homing endonuclease and its host target sequence | | Descriptor: | I-AniI DNA target seq1, I-AniI DNA target seq2, LAGLIDADG endonuclease, ... | | Authors: | Scalley-Kim, M, McConnell Smith, A, Stoddard, B.L. | | Deposit date: | 2007-07-20 | | Release date: | 2008-11-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Coevolution of a homing endonuclease and its host target sequence.
J.Mol.Biol., 372, 2007
|
|
2QTB
 
 | | Human dipeptidyl peptidase iv/cd26 in complex with a 4-aryl cyclohexylalanine inhibitor | | Descriptor: | (2S,3S)-3-AMINO-4-(3,3-DIFLUOROPYRROLIDIN-1-YL)-N,N-DIMETHYL-4-OXO-2-(TRANS-4-[1,2,4]TRIAZOLO[1,5-A]PYRIDIN-6-YLCYCLOHEXYL)BUTANAMIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2007-08-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 4-Arylcyclohexylalanine analogs as potent, selective, and orally active inhibitors of dipeptidyl peptidase IV.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3GMB
 
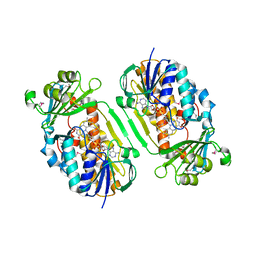 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-03-13 | | Release date: | 2009-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
3WSF
 
 | |
3H57
 
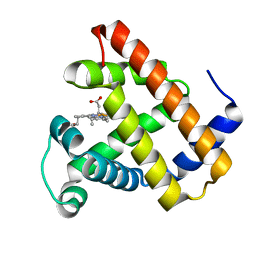 | | Myoglobin Cavity Mutant H64LV68N Deoxy form | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Soman, J, Olson, J.S. | | Deposit date: | 2009-04-21 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optical detection of disordered water within a protein cavity.
J.Am.Chem.Soc., 131, 2009
|
|
2QVB
 
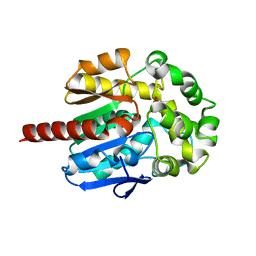 | | Crystal Structure of Haloalkane Dehalogenase Rv2579 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase 3 | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-08 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray crystal structure of Mycobacterium tuberculosis haloalkane dehalogenase Rv2579.
Biochim.Biophys.Acta, 1784, 2008
|
|
2QT1
 
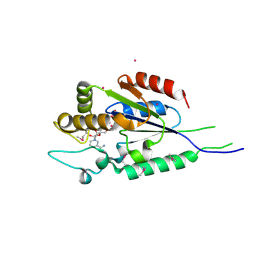 | | Human nicotinamide riboside kinase 1 in complex with nicotinamide riboside | | Descriptor: | Nicotinamide riboside, Nicotinamide riboside kinase 1, PHOSPHATE ION, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
2QTH
 
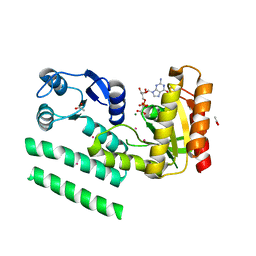 | | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus in complex with GDP | | Descriptor: | ACETATE ION, CADMIUM ION, GTP-binding protein, ... | | Authors: | Wu, H, Sun, L, Brouns, S.J, Fu, S, Rao, Z, Van der Oost, J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus.
To be Published
|
|
3GO8
 
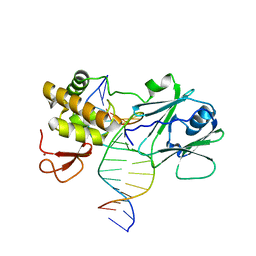 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC3-loop deletion complex | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*(8OG)P*GP*AP*TP*CP*TP*AP*C)-3', 5'-D(P*GP*GP*TP*AP*GP*AP*TP*CP*CP*GP*GP*AP*CP*G)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Spong, M.C, Qi, Y, Verdine, G.L. | | Deposit date: | 2009-03-18 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme
Nature, 462, 2009
|
|
2QU7
 
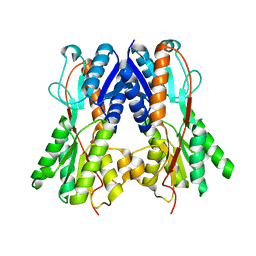 | | Crystal structure of a putative transcription regulator from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-03 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative transcription regulator from Staphylococcus saprophyticus subsp. saprophyticus.
To be Published
|
|
3TVO
 
 | |
2R02
 
 | |
2R0T
 
 | |
