1W55
 
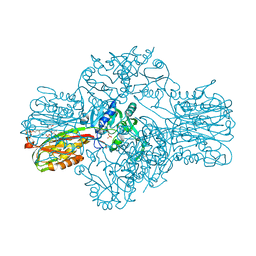 | | Structure of the Bifunctional IspDF from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-MONOPHOSPHATE, GERANYL DIPHOSPHATE, ... | | Authors: | Gabrielsen, M, Bond, C.S, Hunter, W.N. | | Deposit date: | 2004-08-05 | | Release date: | 2004-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hexameric Assembly of the Bifunctional Methylerythritol 2,4-Cyclodiphosphate Synthase and Protein-Protein Associations in the Deoxy-Xylulose-Dependent Pathway of Isoprenoid Precursor Biosynthesis
J.Biol.Chem., 279, 2004
|
|
2O2C
 
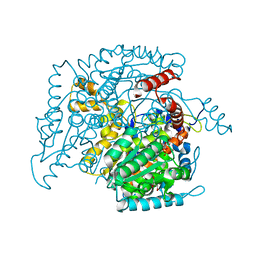 | | Crystal structure of phosphoglucose isomerase from T. brucei containing glucose-6-phosphate in the active site | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Arsenieva, D, Mazock, G.H, Appavu, B.L, Jeffery, C.J. | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of phosphoglucose isomerase from Trypanosoma brucei complexed with glucose-6-phosphate at 1.6 A resolution
Proteins, 74, 2008
|
|
3FE3
 
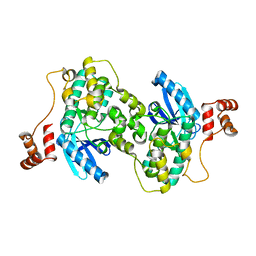 | |
1RKN
 
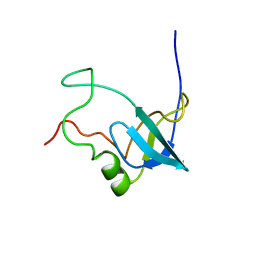 | | Solution structure of 1-110 fragment of Staphylococcal Nuclease with G88W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D.S, Feng, Y.G, Ye, K.Q, Shan, L, Wang, J.F. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
3R8A
 
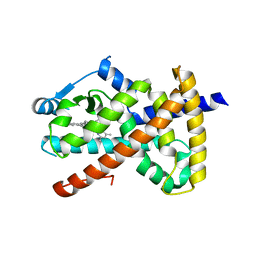 | |
4APT
 
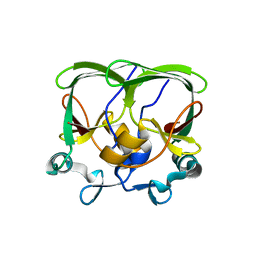 | | The structure of the AXH domain of ataxin-1. | | Descriptor: | ATAXIN-1, SODIUM ION | | Authors: | Rees, M, Chen, Y.W, de Chiara, C, Pastore, A. | | Deposit date: | 2012-04-05 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Self-Assembly and Conformational Heterogeneity of the Axh Domain of Ataxin-1: An Unusual Example of a Chameleon Fold
Biophys.J., 104, 2013
|
|
1V0N
 
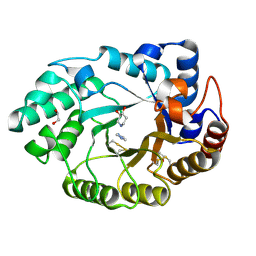 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-isofagomine at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
4AMY
 
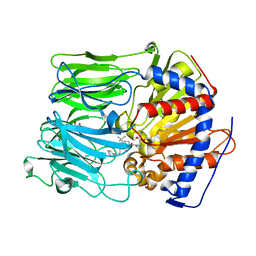 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-1 | | Descriptor: | 1-[(2R,5S)-2-tert-butyl-5-({(2S)-2-[(1R)-1,2-dihydroxyethyl]pyrrolidin-1-yl}carbonyl)pyrrolidin-1-yl]-4-phenylbutan-1-one, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Dynamics, Crystallography and Mutagenesis Studies on the Substrate Gating Mechanism of Prolyl Oligopeptidase.
Biochimie, 94, 2012
|
|
6T9S
 
 | | Human Butyrylcholinesterase in complex with 2-(N-hydroxyimino)-N-[(1S)-3-{4-[(2-methyl-1H-imidazol-1-yl)methyl]-1H-1,2,3-triazol-1-yl}-1- phenylpropyl]acetamide | | Descriptor: | (2~{E})-2-hydroxyimino-~{N}-[(1~{S})-3-[4-[(2-methylimidazol-1-yl)methyl]-1,2,3-triazol-1-yl]-1-phenyl-propyl]ethanamid e, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(5-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Sinko, G, Marakovic, N, Knezevic, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enantioseparation, in vitro testing, and structural characterization of triple-binding reactivators of organophosphate-inhibited cholinesterases.
Biochem.J., 477, 2020
|
|
1V03
 
 | | Crystal structure of the Sorghum bicolor dhurrinase 1 | | Descriptor: | ACETONITRILE, DHURRINASE, PHENOL, ... | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
2UVM
 
 | | Structure of PKBalpha PH domain in complex with a novel inositol headgroup surrogate, benzene 1,2,3,4-tetrakisphosphate | | Descriptor: | BENZENE-1,2,3,4-TETRAYL TETRAKIS[DIHYDROGEN (PHOSPHATE)], RAC-ALPHA SERINE/THREONINE-PROTEIN KINASE, SODIUM ION | | Authors: | Komander, D, Mills, S.J, Trusselle, M.N, Safrany, S.T, van Aalten, D.M.F, Potter, B.V.L. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Inositol Phospholipid Headgroup Surrogate Crystallised in the Pleckstrin Homology Domain of Protein Kinase Balpha.
Acs Chem.Biol., 2, 2007
|
|
1C8K
 
 | | FLAVOPIRIDOL INHIBITS GLYCOGEN PHOSPHORYLASE BY BINDING AT THE INHIBITOR SITE | | Descriptor: | 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Tsitsanou, K.E, Johnson, L.N. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Flavopiridol inhibits glycogen phosphorylase by binding at the inhibitor site.
J.Biol.Chem., 275, 2000
|
|
1RDQ
 
 | | Hydrolysis of ATP in the crystal of Y204A mutant of cAMP-dependent protein kinase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Ten Eyck, L.F, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-05 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a cAMP-dependent Protein Kinase Mutant at 1.26A: New Insights into the Catalytic Mechanism.
J.Mol.Biol., 336, 2004
|
|
1C8S
 
 | | BACTERIORHODOPSIN D96N LATE M STATE INTERMEDIATE | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN ("M" STATE INTERMEDIATE), ... | | Authors: | Luecke, H. | | Deposit date: | 1999-07-29 | | Release date: | 1999-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution.
Science, 286, 1999
|
|
1UZJ
 
 | | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, holo form. | | Descriptor: | CALCIUM ION, FIBRILLIN-1 | | Authors: | Lee, S.S.J, Knott, V, Harlos, K, Handford, P.A, Stuart, D.I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Integrin Binding Fragment from Fibrillin-1 Gives New Insights Into Microfibril Organization
Structure, 12, 2004
|
|
5O42
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal 2,6-pyridinketone inhibitor. | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, 2-fluoranyl-3-[6-[1-(4-fluoranyl-3-oxidanyl-phenyl)ethenyl]pyridin-2-yl]phenol, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Heine, A, Braun, F, Klebe, G, Marchais-Oberwinkler, S. | | Deposit date: | 2017-05-25 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure-based design and profiling of novel 17 beta-HSD14 inhibitors.
Eur J Med Chem, 155, 2018
|
|
1QAZ
 
 | |
4HDO
 
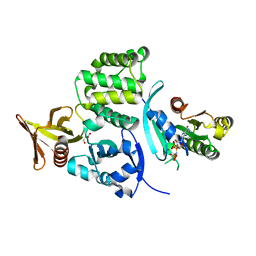 | | Crystal structure of the binary Complex of KRIT1 bound to the Rap1 GTPase | | Descriptor: | GLYCEROL, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Gingras, A.R. | | Deposit date: | 2012-10-02 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Structure of the Ternary Complex of Krev Interaction Trapped 1 (KRIT1) Bound to Both the Rap1 GTPase and the Heart of Glass (HEG1) Cytoplasmic Tail.
J.Biol.Chem., 288, 2013
|
|
4BK6
 
 | |
2F2B
 
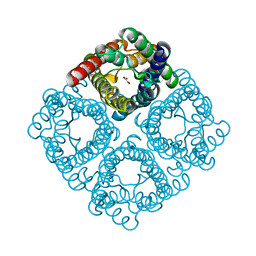 | | Crystal structure of integral membrane protein Aquaporin AqpM at 1.68A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-11-15 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2F3V
 
 | | Solution structure of 1-110 fragment of staphylococcal nuclease with V66W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
1VPC
 
 | | C-TERMINAL DOMAIN (52-96) OF THE HIV-1 REGULATORY PROTEIN VPR, NMR, 1 STRUCTURE | | Descriptor: | VPR PROTEIN | | Authors: | Schueler, W, De Rocquigny, H, Baudat, Y, Sire, J, Roques, B.P. | | Deposit date: | 1998-02-20 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the (52-96) C-terminal domain of the HIV-1 regulatory protein Vpr: molecular insights into its biological functions.
J.Mol.Biol., 285, 1999
|
|
3DCR
 
 | | X-ray structure of HIV-1 protease and hydrated form of ketomethylene isostere inhibitor | | Descriptor: | Chemical analogue HIV-1 protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1QBS
 
 | | HIV-1 PROTEASE INHIBITORS WIIH LOW NANOMOLAR POTENCY | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-25 | | Release date: | 1997-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic HIV protease inhibitors: synthesis, conformational analysis, P2/P2' structure-activity relationship, and molecular recognition of cyclic ureas.
J.Med.Chem., 39, 1996
|
|
3RSX
 
 | |
