4PBT
 
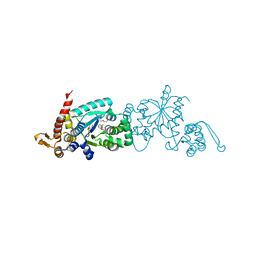 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-trans-cyclooctene-amidopheylalanine (Tco-amF) | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 4-{[(1R,4E)-cyclooct-4-en-1-ylcarbonyl]amino}-L-phenylalanine, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
4PBS
 
 | |
1RKJ
 
 | | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target | | Descriptor: | 5'-R(*GP*GP*AP*UP*GP*CP*CP*UP*CP*CP*CP*GP*AP*GP*UP*GP*CP*AP*UP*CP*C)-3', Nucleolin | | Authors: | Johansson, C, Finger, L.D, Trantirek, L, Mueller, T.D, Kim, S, Laird-Offringa, I.A, Feigon, J. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target.
J.Mol.Biol., 337, 2004
|
|
2AP0
 
 | |
2AP5
 
 | |
4PBR
 
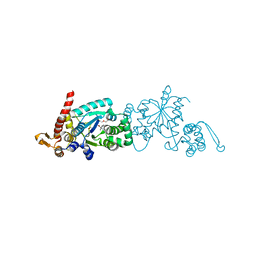 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-(2-bromoisobutyramido)-phenylalanine (BibaF) | | Descriptor: | 4-[(2-bromo-2-methylpropanoyl)amino]-L-phenylalanine, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
4CNF
 
 | | Crystal structure of Sulfolobus acidocaldarius TrmJ | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Van Laer, B, Somme, J, Roovers, M, Steyaert, J, Droogmans, L, Versees, W. | | Deposit date: | 2014-01-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of Two Homologous 2'-O-Methyltransferases Showing Different Specificities for Their tRNA Substrates.
RNA, 20, 2014
|
|
4CNG
 
 | | Crystal structure of Sulfolobus acidocaldarius TrmJ in complex with S-adenosyl-L-Homocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, SPOU RRNA METHYLASE | | Authors: | Van Laer, B, Somme, J, Roovers, M, Steyaert, J, Droogmans, L, Versees, W. | | Deposit date: | 2014-01-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Characterization of Two Homologous 2'-O-Methyltransferases Showing Different Specificities for Their tRNA Substrates.
RNA, 20, 2014
|
|
4HK4
 
 | | Crystal structure of apo Tyrosine-tRNA ligase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystal structure of apo Tyrosine-tRNA ligase mutant protein
To be Published
|
|
4HPW
 
 | | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine | | Descriptor: | 3-methoxy-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine
To be Published
|
|
7E8O
 
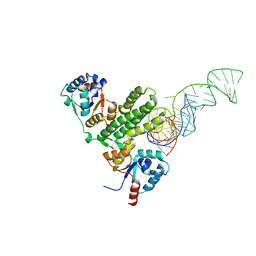 | |
1CQ5
 
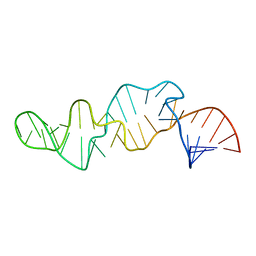 | | NMR STRUCTURE OF SRP RNA DOMAIN IV | | Descriptor: | SRP RNA DOMAIN IV | | Authors: | Schmitz, U, James, T.L, Behrens, S, Freymann, D.M, Lukavsky, P, Walter, P. | | Deposit date: | 1999-08-05 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the phylogenetically most conserved domain of SRP RNA.
RNA, 5, 1999
|
|
1CQL
 
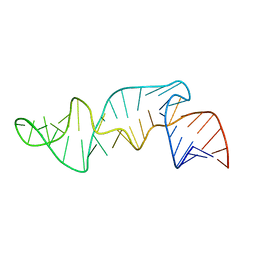 | |
7CHD
 
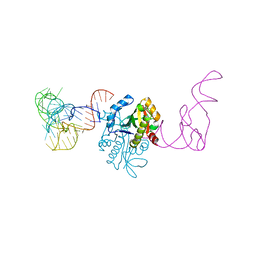 | | AtaT complexed with acetyl-methionyl-tRNAfMet | | Descriptor: | N-acetyltransferase domain-containing protein, RNA (77-MER) | | Authors: | Yashiro, Y, Tomita, K. | | Deposit date: | 2020-07-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.804 Å) | | Cite: | Mechanism of aminoacyl-tRNA acetylation by an aminoacyl-tRNA acetyltransferase AtaT from enterohemorrhagic E. coli.
Nat Commun, 11, 2020
|
|
5WLH
 
 | |
2ZP1
 
 | |
1A1T
 
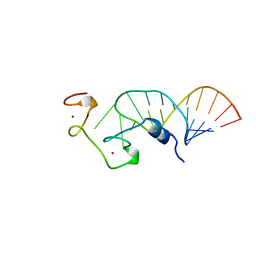 | | STRUCTURE OF THE HIV-1 NUCLEOCAPSID PROTEIN BOUND TO THE SL3 PSI-RNA RECOGNITION ELEMENT, NMR, 25 STRUCTURES | | Descriptor: | NUCLEOCAPSID PROTEIN, SL3 STEM-LOOP RNA, ZINC ION | | Authors: | De Guzman, R.N, Wu, Z.R, Stalling, C.C, Pappalardo, L, Borer, P.N, Summers, M.F. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element.
Science, 279, 1998
|
|
1C2Q
 
 | |
6HMF
 
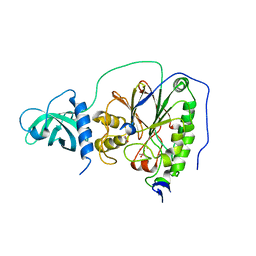 | | D-family DNA polymerase - DP1 subunit (3'-5' proof-reading exonuclease) H451 proof-reading deficient variant | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Raia, P, Delarue, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
6HMS
 
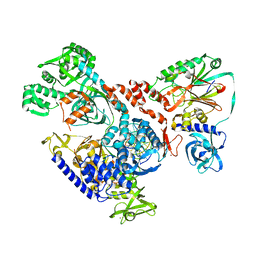 | | Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3'), DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3'), DNA polymerase II large subunit,DNA polymerase II large subunit, ... | | Authors: | Raia, P, Carroni, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
1F78
 
 | |
1F79
 
 | |
1F7H
 
 | |
1F7I
 
 | |
4LWQ
 
 | | Crystal structure of native peptidyl t-RNA hydrolase from Acinetobacter baumannii at 1.38A resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-28 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of native peptidyl t-RNA hydrolase from Acinetobacter baumannii at 1.38A resolution
To be Published
|
|
