7VL3
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with phenyl alpha-D-glucoside | | Descriptor: | (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-phenoxy-oxane-3,4,5-triol, CALCIUM ION, beta-1,2-glucosyltransferase | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
6L4N
 
 | |
7N3E
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C032 | | Descriptor: | C032 Fab Heavy Chain, C032 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7VL6
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with arbutin | | Descriptor: | (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-(4-oxidanylphenoxy)oxane-3,4,5-triol, CALCIUM ION, beta-1,2-glucosyltransferase | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
6L4I
 
 | |
5K68
 
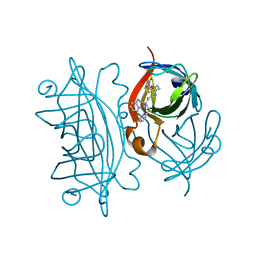 | | Designed Artificial Cupredoxins | | Descriptor: | Streptavidin, [CuII(biot-bu-dpea)]2+ | | Authors: | Mann, S.I, Heinisch, T, Weitz, A.C, Hendrich, M.R, Ward, T.R, Borovik, A.S. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Modular Artificial Cupredoxins.
J.Am.Chem.Soc., 138, 2016
|
|
6LAY
 
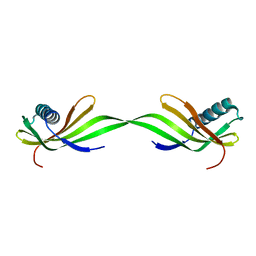 | |
5K6M
 
 | |
5DSG
 
 | | Structure of the M4 muscarinic acetylcholine receptor (M4-mT4L) bound to tiotropium | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Thal, D.M, Kobilka, B.K, Sexton, P.M, Christopoulos, A. | | Deposit date: | 2015-09-17 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the M1 and M4 muscarinic acetylcholine receptors.
Nature, 531, 2016
|
|
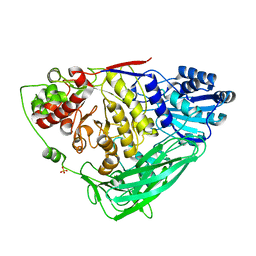
7N3G
 
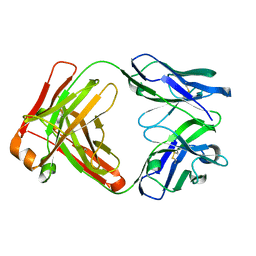 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C098 | | Descriptor: | C098 Fab Heavy Chain, C098 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7VDR
 
 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 13 | | Descriptor: | (1R)-1-[7-[(2-fluoranyl-4-pyrazol-1-yl-phenyl)amino]-1,6-naphthyridin-2-yl]-1-(1-methylpiperidin-4-yl)ethanol, 1,2-ETHANEDIOL, Cyclin-dependent kinase 5 activator 1, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
5K71
 
 | | apo Dbr1 | | Descriptor: | RNA lariat debranching enzyme, putative, SULFATE ION | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The RNA lariat debranching enzyme Dbr1: metal dependence and branched RNA co-crystal structures
Proc.Natl.Acad.Sci.USA, 2016
|
|
7VDP
 
 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclin-dependent kinase 5 activator 1, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
5K0W
 
 | | Crystal structure of the metallo-beta-lactamase GOB-18 from Elizabethkingia meningoseptica | | Descriptor: | CHLORIDE ION, Class B carbapenemase GOB-18, GLYCEROL, ... | | Authors: | Buschiazzo, A, Larrieux, N, Vila, A.J, Lisa, M.N, Moran-Barrio, J. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of the Metallo-beta-Lactamase GOB in the Periplasmic Dizinc Form Reveals an Unusual Metal Site.
Antimicrob.Agents Chemother., 60, 2016
|
|
7PED
 
 | | DEPTOR DEP domain tandem (DEPt) | | Descriptor: | DEP domain-containing mTOR-interacting protein | | Authors: | Waelchli, M, Jakob, R, Maier, T. | | Deposit date: | 2021-08-09 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Regulation of human mTOR complexes by DEPTOR.
Elife, 10, 2021
|
|
5UMZ
 
 | |
7VL0
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with p-nitrophenyl-alpha-D-glucopyranoside | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, Beta-galactosidase, CALCIUM ION | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7N0Z
 
 | |
5UNX
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-(2-(methylamino)ethyl)benzonitrile | | Descriptor: | 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[2-(methylamino)ethyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5E7W
 
 | | X-ray Structure of Human Recombinant 2Zn insulin at 0.92 Angstrom | | Descriptor: | ACETATE ION, Insulin, N-PROPANOL, ... | | Authors: | Lisgarten, D.R, Naylor, C.E, Palmer, R.A, Lobley, C.M.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.9519 Å) | | Cite: | Ultra-high resolution X-ray structures of two forms of human recombinant insulin at 100 K.
Chem Cent J, 11, 2017
|
|
8JNU
 
 | |
5UOD
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 3-[(2-Amino-4-methylquinolin-7-yl)methoxy]-5-((methylamino)methyl)benzonitrile | | Descriptor: | 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(methylamino)methyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
7N3F
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C080 | | Descriptor: | C080 Fab Heavy Chain, C080 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7NAA
 
 | | Crystal structure of Mycobacterium tuberculosis H37Rv PknF kinase domain | | Descriptor: | (4-{[4-(1-benzothiophen-2-yl)pyrimidin-2-yl]amino}phenyl)[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone, Non-specific serine/threonine protein kinase | | Authors: | Oliveira, A.A, Cabarca, S, dos Reis, C.V, Takarada, J.E, Counago, R.M, Balan, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the Mycobacterium tuberculosis c PknF and conformational changes induced in forkhead-associated regulatory domains.
Curr Res Struct Biol, 3, 2021
|
|
5DUU
 
 | |
