2J79
 
 | | Beta-glucosidase from Thermotoga maritima in complex with galacto- hydroximolactam | | Descriptor: | (2E,3R,4R,5R,6S)-3,4,5-TRIHYDROXY-6-(HYDROXYMETHYL)-2-PIPERIDINONE, ACETATE ION, BETA-GLUCOSIDASE A, ... | | Authors: | Gloster, T.M, Zechel, D, Vasella, A, Davies, G.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
2J7E
 
 | | Beta-glucosidase from Thermotoga maritima in complex with methyl acetate-substituted glucoimidazole | | Descriptor: | ACETATE ION, BETA-GLUCOSIDASE A, CALCIUM ION, ... | | Authors: | Gloster, T.M, Zechel, D, Vasella, A, Davies, G.J. | | Deposit date: | 2006-10-07 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
2J7H
 
 | | Beta-glucosidase from Thermotoga maritima in complex with azafagomine | | Descriptor: | ACETATE ION, AZAFAGOMINE, BETA-GLUCOSIDASE A, ... | | Authors: | Gloster, T.M, Meloncelli, P, Stick, R.V, Zechel, D, Davies, G.J. | | Deposit date: | 2006-10-08 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
4KC8
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glycoside hydrolase, ... | | Authors: | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
4KC7
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Glycoside hydrolase, ... | | Authors: | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
5F3D
 
 | | Structure of quinolinate synthase in complex with reaction intermediate W | | Descriptor: | 2-IMINO,3-CARBOXY,5-OXO,6-HYDROXY HEXANOIC ACID, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-12-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Quinolinate Synthase in Complex with a Substrate Analogue, the Condensation Intermediate, and Substrate-Derived Product.
J.Am.Chem.Soc., 138, 2016
|
|
7F3O
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and TAK-653 | | Descriptor: | 7-(4-cyclohexyloxyphenyl)-9-methyl-4$l^{6}-thia-1$l^{4},5,8-triazabicyclo[4.4.0]deca-1(10),6,8-triene 4,4-dioxide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2021-06-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Strictly regulated agonist-dependent activation of AMPA-R is the key characteristic of TAK-653 for robust synaptic responses and cognitive improvement.
Sci Rep, 11, 2021
|
|
3L0O
 
 | | Structure of RNA-free Rho transcription termination factor from Thermotoga maritima | | Descriptor: | SODIUM ION, SULFATE ION, Transcription termination factor rho, ... | | Authors: | Canals, A, Uson, I, Coll, M. | | Deposit date: | 2009-12-10 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structure of RNA-Free Rho Termination Factor Indicates a Dynamic Mechanism of Transcript Capture
J.Mol.Biol., 400, 2010
|
|
7EOB
 
 | | Crystal structure of KIF1A Motor-Neck domain E239K mutant with ADP-Mg-AlFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Kinesin-like protein KIF1A, ... | | Authors: | Ogawa, T, Jiang, X, Hirokawa, N. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A neuropathy-associated kinesin KIF1A mutation hyper-stabilizes the motor-neck interaction during the ATPase cycle.
Embo J., 41, 2022
|
|
7EO9
 
 | | Crystal structure of KIF1A Motor-Neck domain with ADP-Mg-AlFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Kinesin-like protein KIF1A, ... | | Authors: | Ogawa, T, Jiang, X, Hirokawa, N. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A neuropathy-associated kinesin KIF1A mutation hyper-stabilizes the motor-neck interaction during the ATPase cycle.
Embo J., 41, 2022
|
|
5JW7
 
 | | Crystal structure of SopA-Trim56 complex | | Descriptor: | E3 ubiquitin-protein ligase SopA, E3 ubiquitin-protein ligase TRIM56, ZINC ION | | Authors: | Bhogaraju, S, Dikic, I. | | Deposit date: | 2016-05-11 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Structural basis for the recognition and degradation of host TRIM proteins by Salmonella effector SopA.
Nat Commun, 8, 2017
|
|
3G67
 
 | |
5NE6
 
 | |
7F4C
 
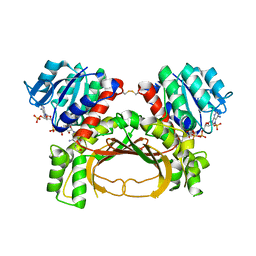 | | The crystal structure of the immature holo-enzyme of homoserine dehydrogenase complexed with NADP and 1,4-butandiol from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | 1,4-BUTANEDIOL, Homoserine dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogata, K, Kaneko, R, Kubota, T, Watanabe, K, Kurihara, E, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
3JUX
 
 | |
5FDF
 
 | |
6BWV
 
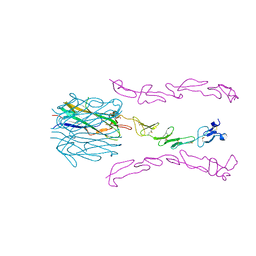 | | Crystal Structure of the 4-1BB/4-1BBL Complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Oganesyan, V, Gilbreth, R.N, Baca, M. | | Deposit date: | 2017-12-15 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human 4-1BB/4-1BBL complex.
J. Biol. Chem., 293, 2018
|
|
3G6B
 
 | |
4YVZ
 
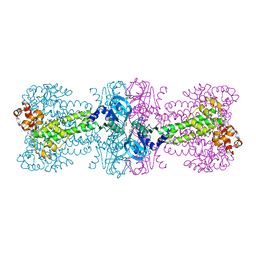 | | Structure of Thermotoga maritima DisA in complex with 3'-dATP/Mn2+ | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, DNA integrity scanning protein DisA, MANGANESE (II) ION | | Authors: | Mueller, M, Deimling, T, Hopfner, K.-P, Witte, G. | | Deposit date: | 2015-03-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structural analysis of the diadenylate cyclase reaction of DNA-integrity scanning protein A (DisA) and its inhibition by 3'-dATP.
Biochem.J., 469, 2015
|
|
4YXM
 
 | | Structure of Thermotoga maritima DisA D75N mutant with reaction product c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA integrity scanning protein DisA | | Authors: | Mueller, M, Deimling, T, Hopfner, K.-P, Witte, G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of the diadenylate cyclase reaction of DNA-integrity scanning protein A (DisA) and its inhibition by 3'-dATP.
Biochem.J., 469, 2015
|
|
6NW5
 
 | |
3DKT
 
 | | Crystal structure of Thermotoga maritima encapsulin | | Descriptor: | Maritimacin, Putative uncharacterized protein | | Authors: | Sutter, M, Boehringer, D, Gutmann, S, Weber-Ban, E, Ban, N. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis of enzyme encapsulation into a bacterial nanocompartment
Nat.Struct.Mol.Biol., 15, 2008
|
|
1OIN
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
4YXJ
 
 | | Structure of Thermotoga maritima DisA in complex with ApCpp | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA integrity scanning protein DisA | | Authors: | Mueller, M, Deimling, T, Hopfner, K.-P, Witte, G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural analysis of the diadenylate cyclase reaction of DNA-integrity scanning protein A (DisA) and its inhibition by 3'-dATP.
Biochem.J., 469, 2015
|
|
4BUB
 
 | | CRYSTAL STRUCTURE OF MURE LIGASE FROM THERMOTOGA MARITIMA IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--LD-LYSINE LIGASE | | Authors: | Favini-Stabile, S, Contreras-Martel, C, Thielens, N, Dessen, A. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mreb and Murg as Scaffolds for the Cytoplasmic Steps of Peptidoglycan Biosynthesis
Environ.Microbiol., 15, 2013
|
|
