6MIC
 
 | |
6MIK
 
 | | Crystal structure of host-guest complex with PP hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
8U5G
 
 | | Crystal structure of the co-expressed SDS22:PP1:I3 complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-09-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The SDS22:PP1:I3 complex: SDS22 binding to PP1 loosens the active site metal to prime metal exchange.
J.Biol.Chem., 300, 2023
|
|
7MO1
 
 | | Crystal Structure of the ZnF1 of Nucleoporin NUP153 in complex with Ran-GDP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
8USE
 
 | |
8QT8
 
 | | Crystal structure of human Sirt2 in complex with a TNFa-Myr analogue TNFn-34 | | Descriptor: | 3-dodecylsulfanylpropanoic acid, NAD-dependent protein deacetylase sirtuin-2, Peptide-based TNFa-Myr analogue TNFn-34, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
7MP4
 
 | | Crystal structure of Epiphyas postvittana antennal carboxylesterase 24 | | Descriptor: | Carboxylesterase-24, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hamiaux, C, Carraher, C. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of an antennally-expressed carboxylesterase suggests lepidopteran odorant degrading enzymes are broadly tuned
Curr Res Insect Sci, 3, 2023
|
|
5TXY
 
 | | Identification of a New Zinc Binding Chemotype of by Fragment Screening on human carbonic anhydrase | | Descriptor: | (5R)-5-phenyl-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, FORMIC ACID, ... | | Authors: | Ren, B, Peat, T.S, Poulsen, S.-A. | | Deposit date: | 2016-11-17 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.206 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
8UZM
 
 | |
5TYA
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-phenyl-1,3-thiazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-18 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
6MIG
 
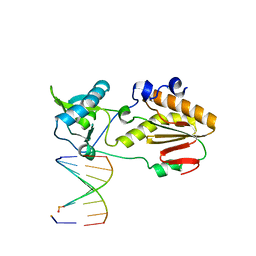 | | Crystal structure of host-guest complex with PB hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), Gag-Pol polyprotein | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
8K8I
 
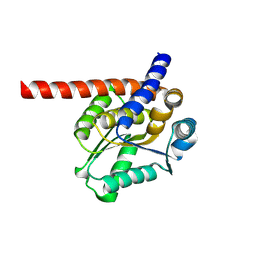 | | De novo design protein -N14 | | Descriptor: | De novo design protein -N14 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo design protein -N14
To Be Published
|
|
5CDE
 
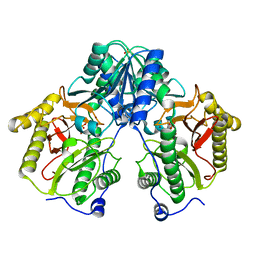 | | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | Proline dipeptidase, SULFATE ION, ZINC ION | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-03 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | R372A mutant of Xaa-Pro dipeptidase from Xanthomonas campestris at 1.85 Angstrom resolution
To Be Published
|
|
8UB6
 
 | |
7OXW
 
 | | CrabP2 mutant R30DK31D | | Descriptor: | ACETATE ION, Cellular retinoic acid-binding protein 2, SULFATE ION | | Authors: | Pastok, M.W, Basle, A, Endicott, J.A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural requirements for the specific binding of CRABP2 to cyclin D3
To Be Published
|
|
8USI
 
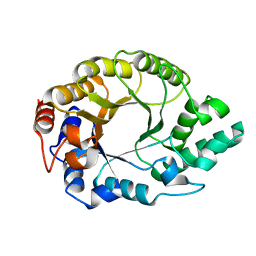 | |
8UZI
 
 | |
7YDL
 
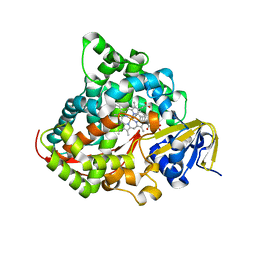 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine
To Be Published
|
|
8K8F
 
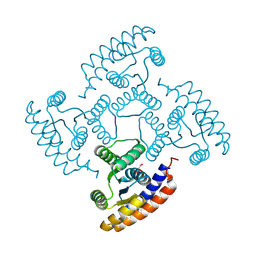 | | De novo design protein -N7 | | Descriptor: | De novo design protein N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo design protein -N7
To Be Published
|
|
5TZB
 
 | | Burkholderia sp. beta-aminopeptidase | | Descriptor: | CALCIUM ION, D-aminopeptidase | | Authors: | McGowan, S, Drinkwater, N, John, M, Dumsday, G. | | Deposit date: | 2016-11-21 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Crystal structure of a beta-aminopeptidase from an Australian Burkholderia sp.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
8V4R
 
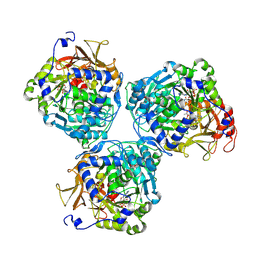 | |
7P4K
 
 | | Soluble epoxide hydrolase in complex with FL217 | | Descriptor: | Bifunctional epoxide hydrolase 2, ~{N}-[[4-(cyclopropylsulfonylamino)-2-(trifluoromethyl)phenyl]methyl]-1-[(3-fluorophenyl)methyl]indole-5-carboxamide | | Authors: | Ni, X, Kramer, J.S, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
5U0C
 
 | |
7MRI
 
 | | Crystal structure of N63T yeast iso-1-cytochrome c | | Descriptor: | Cytochrome c isoform 1, HEME C | | Authors: | Lei, H, Bowler, B.E, Evenson, G.E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Effect on intrinsic peroxidase activity of substituting coevolved residues from Omega-loop C of human cytochrome c into yeast iso-1-cytochrome c.
J.Inorg.Biochem., 232, 2022
|
|
5BX6
 
 | |
