5U0W
 
 | | E. coli dihydropteroate synthase complexed with 9-methylguanine | | Descriptor: | 9-METHYLGUANINE, ACETIC ACID, Dihydropteroate synthase | | Authors: | Chhabra, S, Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | 8-Mercaptoguanine Derivatives as Inhibitors of Dihydropteroate Synthase.
Chemistry, 24, 2018
|
|
7P0I
 
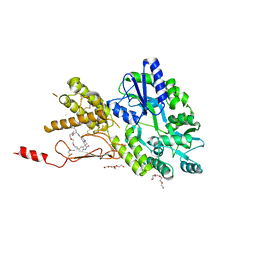 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor Compound 13 | | Descriptor: | (1S,20E)-10-(benzofuran-3-ylmethyl)-12-methyl-15,18-dioxa-5,9,12,24,26-pentazapentacyclo[20.5.2.11,4.13,7.025,28]hentriaconta-3(30),4,6,20,22(29),23,25(28)-heptaene-8,11,27-trione, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Macrocyclic Antagonists of the Calcitonin Gene-Related Peptide Receptor: Design, Realization, and Structural Characterization of Protein-Ligand Complexes.
Acs Chem Neurosci, 13, 2022
|
|
8QF0
 
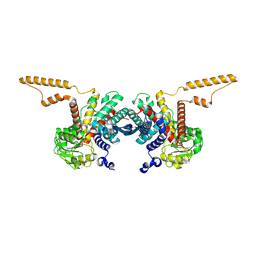 | |
7P0F
 
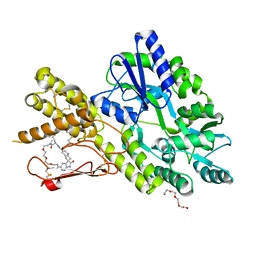 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0028125 | | Descriptor: | (1S,10R,23E)-12-methyl-10-[(7-methyl-1H-indazol-5-yl)methyl]-15,18,21-trioxa-5,9,12,27,29-pentazapentacyclo[23.5.2.11,4.13,7.028,31]tetratriaconta-3(33),4,6,23,25(32),26,28(31)-heptaene-8,11,30-trione, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2021-06-29 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel Macrocyclic Antagonists of the Calcitonin Gene-Related Peptide Receptor: Design, Realization, and Structural Characterization of Protein-Ligand Complexes.
Acs Chem Neurosci, 13, 2022
|
|
7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | Descriptor: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | Authors: | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
5GVS
 
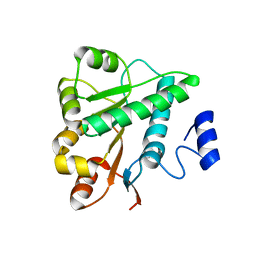 | | Crystal structure of the DDX41 DEAD domain in an apo open form | | Descriptor: | Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
7UYO
 
 | |
7UYN
 
 | |
7UYP
 
 | |
7OQG
 
 | |
5TP3
 
 | |
7MHD
 
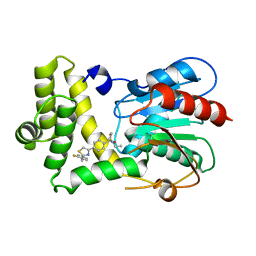 | | Thioesterase Domain of Human Fatty Acid Synthase (FASN-TE) binding a competitive inhibitor SBP-7635 | | Descriptor: | Fatty acid synthase, N,N-diethyl-4-{2-[(2-fluorophenyl)methyl]-1,3-thiazol-4-yl}benzene-1-sulfonamide | | Authors: | Aleshin, A.E, Lambert, L, Liddington, R.C, Cosford, N. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Thioesterase Domain of Human Fatty Acid Synthase (FASN-TE) binding a competitive inhibitor SBP-7635
To Be Published
|
|
8K9S
 
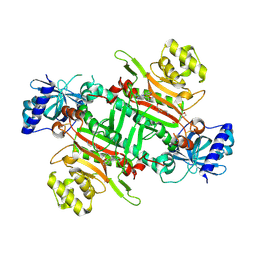 | | Crystal structure of plasmodium LysRS complexing with ASP3026 derived LysRS inhibitor 1 (ADKI1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Zhou, J, Xia, M, Yang, G, Li, P, Fang, P. | | Deposit date: | 2023-08-01 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided conversion from an anaplastic lymphoma kinase inhibitor into Plasmodium lysyl-tRNA synthetase selective inhibitors.
Commun Biol, 7, 2024
|
|
7OQJ
 
 | |
7V1A
 
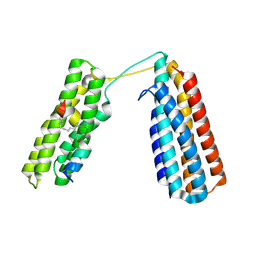 | | Stapled TBS peptide from RIAM bound to talin R7R8 domains | | Descriptor: | 1,2-ETHANEDIOL, ASP-ILE-ASP-GLN-MET-PHE-SER-THR-LEU-LEU-GLY-GLU-MK8-ASP-LEU-LEU-MK8-GLN-SER, Talin-1 | | Authors: | Zhang, P, Gao, T, Wu, J. | | Deposit date: | 2022-05-11 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Inhibition of talin-induced integrin activation by a double-hit stapled peptide.
Structure, 31, 2023
|
|
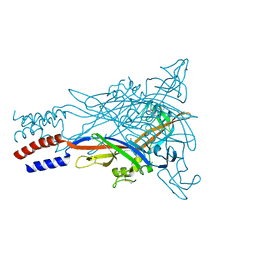
5U2H
 
 | |
7OQS
 
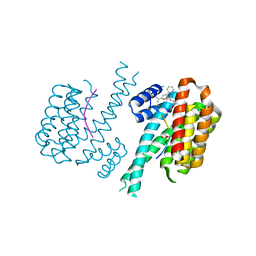 | | Ternary complex of 14-3-3 sigma, Amot-p130 phosphopeptide, and WQ177 | | Descriptor: | 14-3-3 protein sigma, 2-(1,3-benzodioxol-5-yl)-~{N}-[(5-carbamimidoyl-3-phenyl-thiophen-2-yl)methyl]ethanamide, Amot-p130 phosphopeptide (pS175), ... | | Authors: | Centorrino, F, Ottmann, C. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
7XV1
 
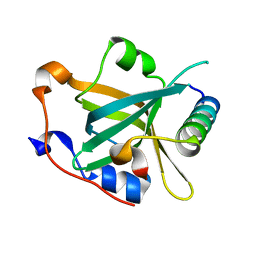 | | Crystal structure of RPA70N-HelB fusion | | Descriptor: | DNA helicase B, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Wu, Y.Y, Zang, N, Fu, W.M, Zhou, C. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
7UXW
 
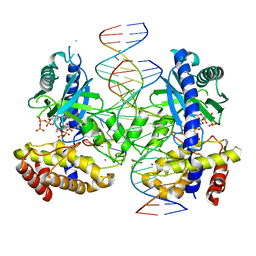 | | Structure of ATP and GTP bind to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7UYQ
 
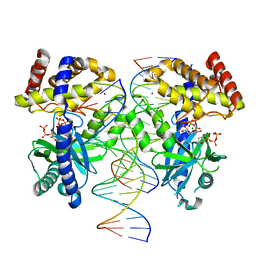 | | Structure of GTP binds to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7XUT
 
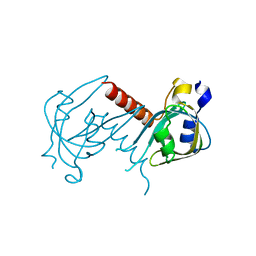 | | Crystal structure of RPA70N-WRN fusion | | Descriptor: | fusion protein of Replication protein A 70 kDa DNA-binding subunit and Werner syndrome ATP-dependent helicase | | Authors: | Wu, Y.Y, Zang, N, Fu, W.M, Zhou, C. | | Deposit date: | 2022-05-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
8JSV
 
 | |
7OR3
 
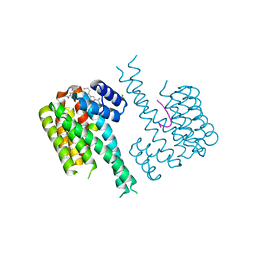 | | Ternary complex of 14-3-3 sigma, NotchpS1917 phosphopeptide, and WQ136 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
5GXH
 
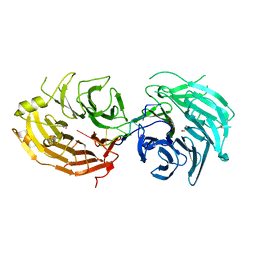 | | The structure of the Gemin5 WD40 domain with AAUUUUUG | | Descriptor: | GLYCEROL, Gem-associated protein 5, RNA (5'-R(*A*AP*UP*UP*UP*UP*UP*G)-3'), ... | | Authors: | Xu, C, He, H, Li, Y, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-17 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly
Genes Dev., 30, 2016
|
|
8UCX
 
 | |
