8EHZ
 
 | | Crystal structure of the STUB1 TPR domain in complex with H317, a Helicon Polypeptide | | Descriptor: | E3 ubiquitin-protein ligase CHIP, H317, N,N'-(1,4-phenylene)diacetamide | | Authors: | Li, K, Swiecicki, J.-M, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
7ORP
 
 | | crystal structure of human carbonic anhydrase II in complex with 4-((2-hydroxy-3-((3,4,5-trimethoxyphenyl)tellanyl)propyl)selanyl)benzenesulfonamide | | Descriptor: | 4-[(2R)-2-oxidanyl-3-(3,4,5-trimethylphenyl)tellanyl-propyl]selanylbenzenesulfonamide, 4-[(2S)-2-oxidanyl-3-(3,4,5-trimethoxyphenyl)tellanyl-propyl]selanylbenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Chalcogenides-incorporating carbonic anhydrase inhibitors concomitantly reverted oxaliplatin-induced neuropathy and enhanced antiproliferative action.
Eur.J.Med.Chem., 225, 2021
|
|
8EI0
 
 | | Crystal structure of the STUB1 TPR domain in complex with H318, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, H318, ... | | Authors: | Li, K, Swiecicki, J.-M, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EI6
 
 | | Crystal structure of the WWP2 HECT domain in complex with H305, a Helicon Polypeptide | | Descriptor: | H305, N,N'-(1,4-phenylene)diacetamide, NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EI5
 
 | | Crystal structure of the WWP2 HECT domain in complex with H301, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, H301, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
7OTE
 
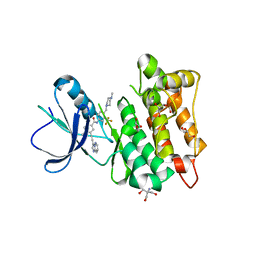 | | Src Kinase Domain in complex with ponatinib | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, GLYCEROL, ... | | Authors: | Soriano-Maldonado, P, Cuesta-Hernandez, H.N, Plaza-Menacho, I. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric switch between the activation loop and a c-terminal palindromic phospho-motif controls c-Src function.
Nat Commun, 14, 2023
|
|
7ORQ
 
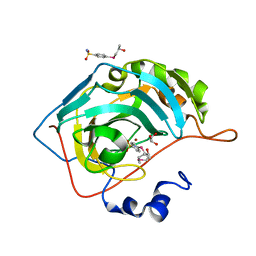 | |
5NQS
 
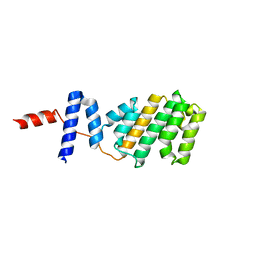 | | Structure of the Arabidopsis Thaliana TOPLESS N-terminal domain | | Descriptor: | Protein TOPLESS | | Authors: | Nanao, M.H, Arevalillo, M.R, Vinos-Poyo, T, Parcy, F, Dumas, R. | | Deposit date: | 2017-04-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6M9L
 
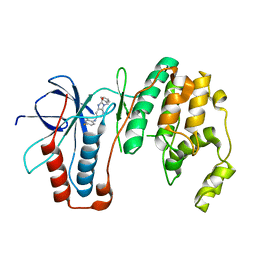 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping - compound 10 | | Descriptor: | 3-benzyl-6-[(2,4-difluorophenyl)amino]-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-23 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
3NSF
 
 | | Apo form of the multicopper oxidase CueO | | Descriptor: | Blue copper oxidase cueO | | Authors: | Roberts, S.A, Montfort, W.R, Singh, S.K. | | Deposit date: | 2010-07-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
3NSC
 
 | | C500S MUTANT OF CueO BOUND TO Cu(II) | | Descriptor: | ACETATE ION, Blue copper oxidase cueO, COPPER (II) ION, ... | | Authors: | Roberts, S.A, Montfort, W.R, Singh, S.K. | | Deposit date: | 2010-07-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
7S7J
 
 | | Structure of Human SPASTIN-IST1 complex. | | Descriptor: | CALCIUM ION, CHLORIDE ION, IST1 homolog, ... | | Authors: | Skalicky, J.J, Sundquist, W.I. | | Deposit date: | 2021-09-16 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Comprehensive analysis of the human ESCRT-III-MIT domain interactome reveals new cofactors for cytokinetic abscission.
Elife, 11, 2022
|
|
6MA6
 
 | | Human CYP3A4 bound to an inhibitor metyrapone | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, GLYCEROL, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-08-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Interaction of Human Drug-Metabolizing CYP3A4 with Small Inhibitory Molecules.
Biochemistry, 58, 2019
|
|
6LLB
 
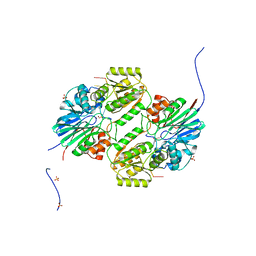 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, in complex with 6 nt RNA | | Descriptor: | MPY-RNase J, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, ... | | Authors: | Li, D.F, Hou, Y.J, Guo, L. | | Deposit date: | 2019-12-22 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A newly identified duplex RNA unwinding activity of archaeal RNase J depends on processive exoribonucleolysis coupled steric occlusion by its structural archaeal loops.
Rna Biol., 17, 2020
|
|
5OVV
 
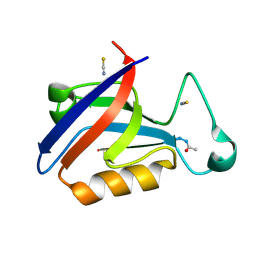 | | PDZ domain from rat Shank3 bound to the C terminus of ProSAPiP1 | | Descriptor: | Leucine zipper putative tumor suppressor 3, SH3 and multiple ankyrin repeat domains protein 3, THIOCYANATE ION | | Authors: | Ponna, S.K, Myllykoski, M, Boeckers, T.M, Kursula, P. | | Deposit date: | 2017-08-29 | | Release date: | 2018-03-07 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for PDZ domain interactions in the post-synaptic density scaffolding protein Shank3.
J. Neurochem., 145, 2018
|
|
3NT0
 
 | |
3QQX
 
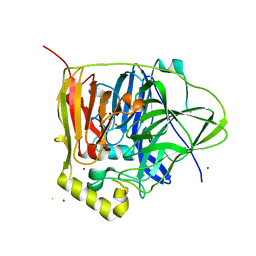 | | Reduced Native Intermediate of the Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (I) ION, COPPER (II) ION, ... | | Authors: | Montfort, W.R, Roberts, S.A, Singh, S.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CueO E506D Mutant: Crystal Structure of Reduced Native Intermediate, Kinetics, and Impairment of Product Release
To be Published
|
|
2DRC
 
 | |
8G0Q
 
 | |
8FFT
 
 | | Structure of GntC, a PLP-dependent enzyme catalyzing L-enduracididine biosynthesis from (S)-4-hydroxy-L-arginine | | Descriptor: | Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, MAGNESIUM ION | | Authors: | Chen, P.Y.-T, Lima, S.T, Chekan, J.R, Moore, B.S. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Acs Catalysis, 13, 2023
|
|
8FCK
 
 | | Structure of the vertebrate augmin complex | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, Green fluorescent protein chimera, HAUS augmin like complex subunit 4 L homeolog, ... | | Authors: | Travis, S.M, Huang, W, Zhang, R, Petry, S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.88 Å) | | Cite: | Integrated model of the vertebrate augmin complex.
Nat Commun, 14, 2023
|
|
8FFU
 
 | | Structure of GntC, a PLP-dependent enzyme catalyzing L-enduracididine biosynthesis from (S)-4-hydroxy-L-arginine, with the substrate bound | | Descriptor: | (2S,4S)-5-carbamimidamido-4-hydroxy-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pentanoic acid (non-preferred name), Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, MAGNESIUM ION | | Authors: | Chen, P.Y.-T, Moore, B.S. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Acs Catalysis, 13, 2023
|
|
8G2E
 
 | | PKM2 bound to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-aminophenyl)methyl]-5-methyl-7-[methyl(oxidanyl)-$l^{3}-sulfanyl]pyridazino[4,5-b]indol-4-one, MAGNESIUM ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Development of Novel Small-Molecule Activators of Pyruvate Kinase Muscle Isozyme 2, PKM2, to Reduce Photoreceptor Apoptosis.
Pharmaceuticals, 16, 2023
|
|
6MA7
 
 | | Human CYP3A4 bound to an inhibitor fluconazole | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Cytochrome P450 3A4, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-08-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Interaction of Human Drug-Metabolizing CYP3A4 with Small Inhibitory Molecules.
Biochemistry, 58, 2019
|
|
7SCT
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001 Complexed to Human Chemokine CCL16 | | Descriptor: | C-C motif chemokine 16, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Engineering broad-spectrum inhibitors of inflammatory chemokines from subclass A3 tick evasins.
Nat Commun, 14, 2023
|
|
