3ZMT
 
 | | LSD1-CoREST in complex with PRSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
6FQQ
 
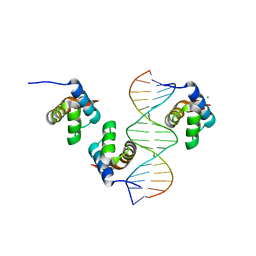 | |
2XTD
 
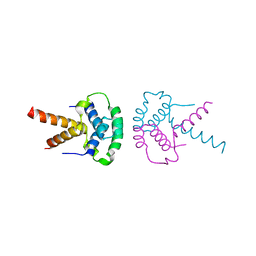 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | TBL1 F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6GZ1
 
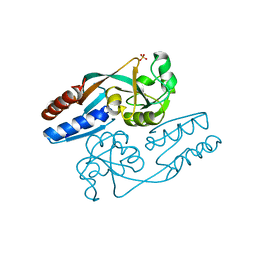 | | Crystal Structure of the LeuO Effector Binding Domain | | Descriptor: | HTH-type transcriptional regulator LeuO, SULFATE ION | | Authors: | Fragel, S, Montada, A.M, Baumann, U, Schacherl, M, Schnetz, K. | | Deposit date: | 2018-07-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Characterization of the pleiotropic LysR-type transcription regulator LeuO of Escherichia coli.
Nucleic Acids Res., 47, 2019
|
|
2VXF
 
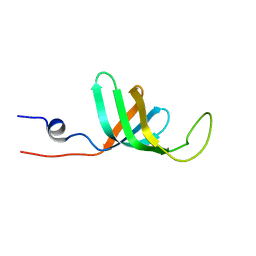 | |
2IZ8
 
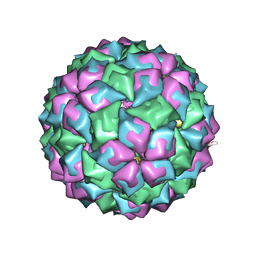 | | MS2-RNA HAIRPIN (C-7) COMPLEX | | Descriptor: | MS2 COAT PROTEIN, RNA, (5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*CP*UP*CP* AP*CP*CP*CP*AP*UP*GP*U)-3') | | Authors: | Helgstrand, C, Grahn, E, Stonehouse, N.J, Stockley, P.G, Liljas, L. | | Deposit date: | 2006-07-25 | | Release date: | 2006-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Investigating the Structural Basis of Purine Specificity in the Structures of MS2 Coat Protein RNA Translational Operator Hairpins
Nucleic Acids Res., 30, 2002
|
|
3PXP
 
 | |
6TBT
 
 | |
6TCJ
 
 | |
3K33
 
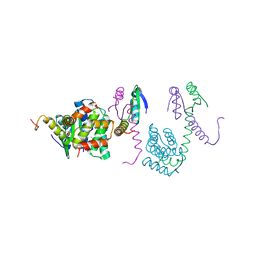 | | Crystal structure of the Phd-Doc complex | | Descriptor: | ACETATE ION, Death on curing protein, Polypeptide of unknown amino acids and source, ... | | Authors: | Loris, R, Garcia-Pino, A. | | Deposit date: | 2009-10-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity.
Cell(Cambridge,Mass.), 142, 2010
|
|
4JG3
 
 | | Crystal structure of catabolite repression control protein (crc) from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Catabolite repression control protein | | Authors: | Grishkovskaya, I, Milojevic, T, Sonnleitner, E, Blaesi, U, Djinovic-Carugo, K. | | Deposit date: | 2013-02-28 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Pseudomonas aeruginosa Catabolite Repression Control Protein Crc Is Devoid of RNA Binding Activity
Plos One, 8, 2013
|
|
3MW6
 
 | | Crystal structure of NMB1681 from Neisseria meningitidis MC58, a FinO-like RNA chaperone | | Descriptor: | GLYCEROL, uncharacterized protein NMB1681 | | Authors: | Tan, K, Zhou, M, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | N. meningitidis 1681 is a member of the FinO family of RNA chaperones.
Rna Biol., 7, 2010
|
|
4LZL
 
 | |
5EY0
 
 | | Crystal structure of CodY from Staphylococcus aureus with GTP and Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, GUANOSINE-5'-TRIPHOSPHATE, ISOLEUCINE | | Authors: | Han, A, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
5X60
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 9 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
5EY1
 
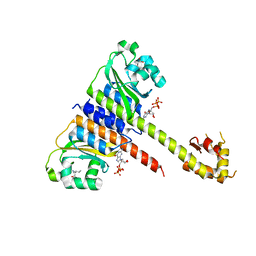 | | Crystal structure of CodY from Staphylococcus aureus with GTP and Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, GUANOSINE-5'-TRIPHOSPHATE, ISOLEUCINE | | Authors: | Han, A, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
7B22
 
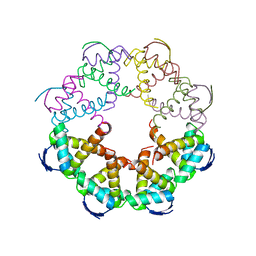 | | Vibrio cholerae ParD2 Antitoxin | | Descriptor: | Antitoxin ParD | | Authors: | Garcia-Rodriguez, G, Loris, R. | | Deposit date: | 2020-11-25 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Entropic pressure controls the oligomerization of the Vibrio cholerae ParD2 antitoxin.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3HTH
 
 | |
3C2I
 
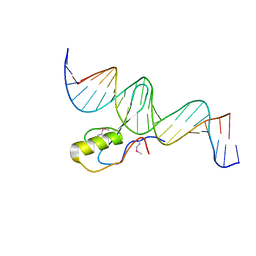 | | The Crystal Structure of Methyl-CpG Binding Domain of Human MeCP2 in Complex with a Methylated DNA Sequence from BDNF | | Descriptor: | DNA (5'-D(*DAP*DTP*DAP*DGP*DAP*DAP*DGP*DAP*DAP*DTP*DTP*DCP*(5CM)P*DGP*DTP*DTP*DCP*DCP*DAP*DG)-3'), DNA (5'-D(*DTP*DCP*DTP*DGP*DGP*DAP*DAP*(5CM)P*DGP*DGP*DAP*DAP*DTP*DTP*DCP*DTP*DTP*DCP*DTP*DA)-3'), Methyl-CpG-binding protein 2 | | Authors: | Ho, K.L, McNae, I.W, Schmiedeberg, L, Klose, R.J, Bird, A.P, Walkinshaw, M.D. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MeCP2 binding to DNA depends upon hydration at methyl-CpG
Mol.Cell, 29, 2008
|
|
3HTA
 
 | |
2W1T
 
 | | Crystal Structure of B. subtilis SpoVT | | Descriptor: | STAGE V SPORULATION PROTEIN T | | Authors: | Asen, I, Djuranovic, S, Lupas, A.N, Zeth, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Spovt, the Final Modulator of Gene Expression During Spore Development in Bacillus Subtilis
J.Mol.Biol., 386, 2009
|
|
3QPH
 
 | | The three-dimensional structure of TrmB, a global transcriptional regulator of the hyperthermophilic archaeon Pyrococcus furiosus in complex with sucrose | | Descriptor: | ACETATE ION, GLYCEROL, TrmB, ... | | Authors: | Krug, M, Lee, S.-J, Boos, W, Welte, W, Diederichs, K. | | Deposit date: | 2011-02-13 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | The three-dimensional structure of TrmB, a transcriptional regulator of dual function in the hyperthermophilic archaeon Pyrococcus furiosus in complex with sucrose.
Protein Sci., 22, 2013
|
|
3HTJ
 
 | |
5U8K
 
 | | RitR mutant - C128S | | Descriptor: | Response regulator | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2016-12-14 | | Release date: | 2017-12-20 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization.
PLoS Pathog., 14, 2018
|
|
2F3X
 
 | |
