3HSF
 
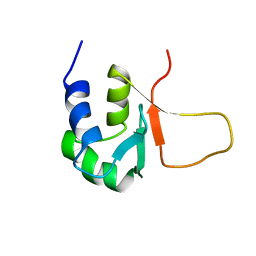 | | HEAT SHOCK TRANSCRIPTION FACTOR (HSF) | | Descriptor: | HEAT SHOCK TRANSCRIPTION FACTOR | | Authors: | Damberger, F.F, Pelton, J.G, Liu, C, Cho, H, Harrison, C.J, Nelson, H.C.M, Wemmer, D.E. | | Deposit date: | 1995-08-07 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure and dynamics of the DNA-binding domain of the heat shock factor from Kluyveromyces lactis.
J.Mol.Biol., 254, 1995
|
|
1TT0
 
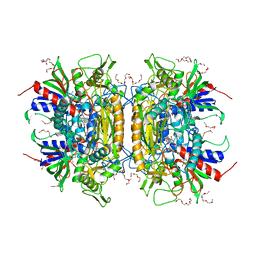 | | Crystal Structure of Pyranose 2-Oxidase | | Descriptor: | ACETATE ION, DODECAETHYLENE GLYCOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hallberg, B.M, Leitner, C, Haltrich, D, Divne, C. | | Deposit date: | 2004-06-21 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the 270 kDa homotetrameric lignin-degrading enzyme pyranose 2-oxidase
J.Mol.Biol., 341, 2004
|
|
1VK5
 
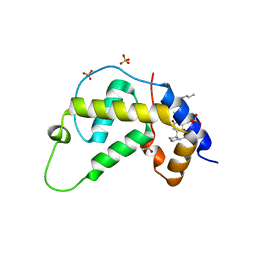 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At3g22680 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, SULFATE ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structure at 1.6 A resolution of the protein from gene locus At3g22680 from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1M70
 
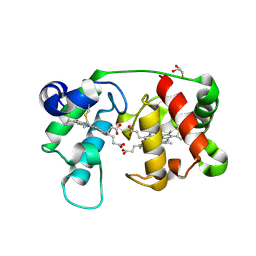 | |
1EGG
 
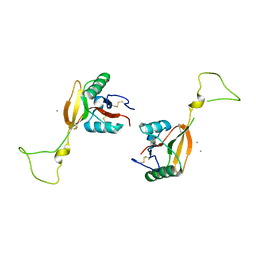 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
2J5B
 
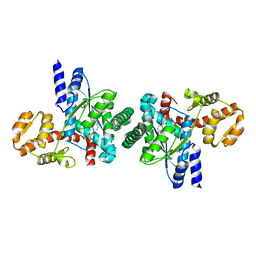 | | Structure of the Tyrosyl tRNA synthetase from Acanthamoeba polyphaga Mimivirus complexed with tyrosynol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, TYROSYL-TRNA SYNTHETASE | | Authors: | Abergel, C, Rudinger-thirion, J, Giege, R, Claverie, J.M. | | Deposit date: | 2006-09-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Virus-Encoded Aminoacyl-tRNA Synthetases: Structural and Functional Characterization of Mimivirus Tyrrs and Metrs.
J.Virol., 81, 2007
|
|
1APL
 
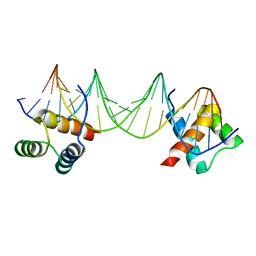 | | CRYSTAL STRUCTURE OF A MAT-ALPHA2 HOMEODOMAIN-OPERATOR COMPLEX SUGGESTS A GENERAL MODEL FOR HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*C P*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT-ALPHA2 HOMEODOMAIN) | | Authors: | Wolberger, C, Vershon, A.K, Liu, B, Johnson, A.D, Pabo, C.O. | | Deposit date: | 1993-10-04 | | Release date: | 1993-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 67, 1991
|
|
1F41
 
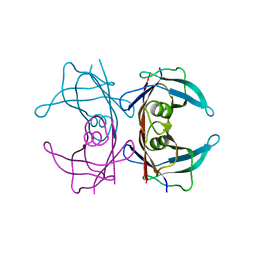 | | CRYSTAL STRUCTURE OF HUMAN TRANSTHYRETIN AT 1.5A RESOLUTION | | Descriptor: | TRANSTHYRETIN | | Authors: | Hornberg, A, Eneqvist, T, Olofsson, A, Lundgren, E, Sauer-Eriksson, A.E. | | Deposit date: | 2000-06-07 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A comparative analysis of 23 structures of the amyloidogenic protein transthyretin.
J.Mol.Biol., 302, 2000
|
|
1FIH
 
 | | N-ACETYLGALACTOSAMINE BINDING MUTANT OF MANNOSE-BINDING PROTEIN A (QPDWG-HDRPY), COMPLEX WITH N-ACETYLGALACTOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Feinberg, H, Torgerson, D, Drickamer, K, Weis, W.I. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of pH-dependent N-acetylgalactosamine binding by a functional mimic of the hepatocyte asialoglycoprotein receptor.
J.Biol.Chem., 275, 2000
|
|
388D
 
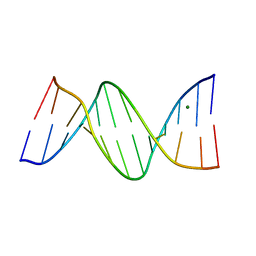 | | CRYSTAL STRUCTURE OF B-DNA WITH INCORPORATED 2'-DEOXY-2'-FLUORO-ARABINO-FURANOSYL THYMINES: IMPLICATIONS OF CONFORMATIONAL PREORGANIZATION FOR DUPLEX STABILITY | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TAF)P*(TAF)P*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Berger, I, Tereshko, V, Ikeda, H, Marquez, V.E, Egli, M. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of B-DNA with incorporated 2'-deoxy-2'-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability.
Nucleic Acids Res., 26, 1998
|
|
1FAO
 
 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM DAPP1/PHISH IN COMPLEX WITH INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | | Descriptor: | DUAL ADAPTOR OF PHOSPHOTYROSINE AND 3-PHOSPHOINOSITIDES, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-07-13 | | Release date: | 2000-07-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
1FIF
 
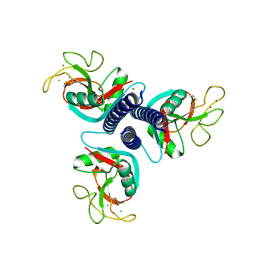 | | N-ACETYLGALACTOSAMINE-SELECTIVE MUTANT OF MANNOSE-BINDING PROTEIN-A (QPDWG-HDRPY) | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN-A | | Authors: | Feinberg, H, Torgersen, D, Drickamer, K, Weis, W.I. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of pH-dependent N-acetylgalactosamine binding by a functional mimic of the hepatocyte asialoglycoprotein receptor.
J.Biol.Chem., 275, 2000
|
|
389D
 
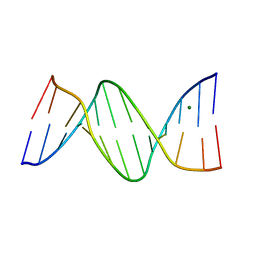 | | CRYSTAL STRUCTURE OF B-DNA WITH INCORPORATED 2'-DEOXY-2'-FLUORO-ARABINO-FURANOSYL THYMINES: IMPLICATIONS OF CONFORMATIONAL PREORGANIZATION FOR DUPLEX STABILITY | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TAF)P*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Berger, I, Tereshko, V, Ikeda, H, Marquez, V.E, Egli, M. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of B-DNA with incorporated 2'-deoxy-2'-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability.
Nucleic Acids Res., 26, 1998
|
|
1FHX
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
1FB8
 
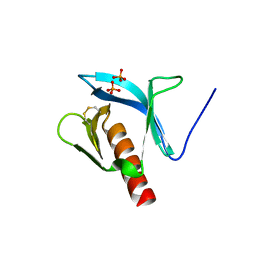 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM DAPP1/PHISH | | Descriptor: | DUAL ADAPTOR OF PHOSPHOTYROSINE AND 3-PHOSPHOINOSITIDES, PHOSPHATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-07-14 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
2BTH
 
 | |
1FHW
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol(1,3,4,5,6)pentakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
2BTG
 
 | |
1FJM
 
 | | Protein serine/threonine phosphatase-1 (alpha isoform, type 1) complexed with microcystin-LR toxin | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, PROTEIN SERINE/THREONINE PHOSPHATASE-1 (ALPHA ISOFORM, ... | | Authors: | Goldberg, J, Nairn, A.C, Kuriyan, J. | | Deposit date: | 1995-12-17 | | Release date: | 1996-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1.
Nature, 376, 1995
|
|
1KTV
 
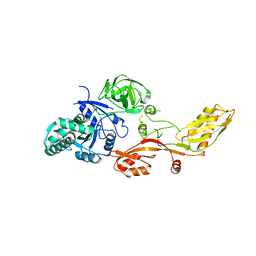 | |
2CMF
 
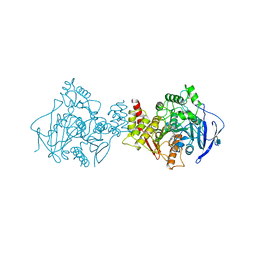 | | Torpedo californica acetylcholinesterase complexed with alkylene- linked bis-tacrine dimer (5 carbon linker) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, N,N'-DI-1,2,3,4-TETRAHYDROACRIDIN-9-YLPENTANE-1,5-DIAMINE | | Authors: | Rydberg, E.H, Brumshtein, B, Greenblatt, H.M, Wong, D.M, Pang, Y.P, Silman, I, Sussman, J.L. | | Deposit date: | 2006-05-08 | | Release date: | 2006-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complexes of alkylene-linked tacrine dimers with Torpedo californica acetylcholinesterase: Binding of Bis5-tacrine produces a dramatic rearrangement in the active-site gorge.
J. Med. Chem., 49, 2006
|
|
8ATG
 
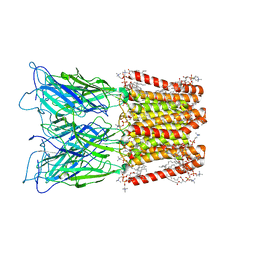 | | Pentameric ligand-gated ion channel GLIC with bound lipids | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Proton-gated ion channel | | Authors: | Bergh, C, Rovsnik, U, Howard, R.J, Lindahl, E. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of lipid binding sites in a ligand-gated ion channel by integrating simulations and cryo-EM.
Elife, 12, 2024
|
|
1GML
 
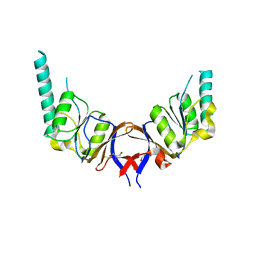 | | crystal structure of the mouse CCT gamma apical domain (triclinic) | | Descriptor: | GLYCEROL, T-COMPLEX PROTEIN 1 SUBUNIT GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-09-17 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
1GN1
 
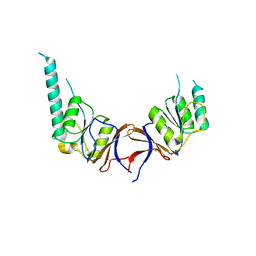 | | crystal structure of the mouse CCT gamma apical domain (monoclinic) | | Descriptor: | CALCIUM ION, CCT-GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-10-01 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
5EDV
 
 | | Structure of the HOIP-RBR/UbcH5B~ubiquitin transfer complex | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Lechtenberg, B.C, Mace, P.D, Sanishvili, R, Riedl, S.J. | | Deposit date: | 2015-10-22 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation.
Nature, 529, 2016
|
|
