7XIE
 
 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*GP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A•G•G•G•G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
7X7G
 
 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
7XDH
 
 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*AP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
2LSG
 
 | |
6FQ2
 
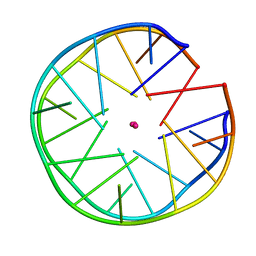 | | Structure of minimal sequence for left -handed G-quadruplex formation | | Descriptor: | DNA (5'-D(*GP*TP*GP*GP*TP*GP*GP*TP*GP*GP*TP*G)-3'), POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Heddi, B, Bakalar, B. | | Deposit date: | 2018-02-13 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6FTU
 
 | | Structure of a Quadruplex forming sequence from D. discoideum | | Descriptor: | DNA (26-MER), POTASSIUM ION | | Authors: | Guedin, A, Linda, L, Armane, S, Lacroix, L, Mergny, J.L, Thore, S, Yatsunyk, L.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Quadruplexes in 'Dicty': crystal structure of a four-quartet G-quadruplex formed by G-rich motif found in the Dictyostelium discoideum genome.
Nucleic Acids Res., 46, 2018
|
|
6GZ6
 
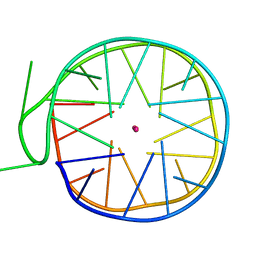 | | Structure of a left-handed G-quadruplex | | Descriptor: | DNA (27-MER), POTASSIUM ION | | Authors: | Bakalar, B, Heddi, B, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2018-07-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4AD8
 
 | | Crystal structure of a deletion mutant of Deinococcus radiodurans RecN | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.998 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
4ABX
 
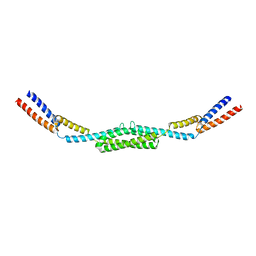 | | Crystal structure of Deinococcus radiodurans RecN coiled-coil domain | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
1RIF
 
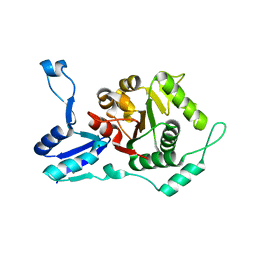 | |
4ABY
 
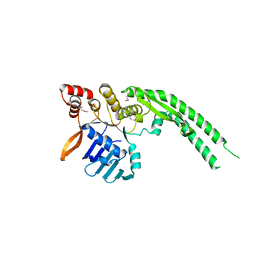 | | Crystal structure of Deinococcus radiodurans RecN head domain | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-12 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
250D
 
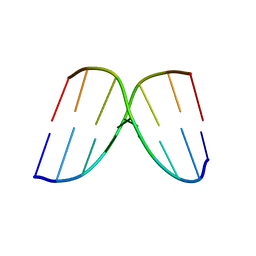 | | STRUCTURAL COMPARISON BETWEEN THE D(CTAG) SEQUENCE IN OLIGONUCLEOTIDES AND TRP AND MET REPRESSOR-OPERATOR COMPLEXES | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3') | | Authors: | Urpi, L, Tereshko, V, Malinina, L, Huynh-Dinh, T, Subirana, J.A. | | Deposit date: | 1996-02-22 | | Release date: | 1996-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural comparison between the d(CTAG) sequence in oligonucleotides and trp and met repressor-operator complexes.
Nat.Struct.Biol., 3, 1996
|
|
176D
 
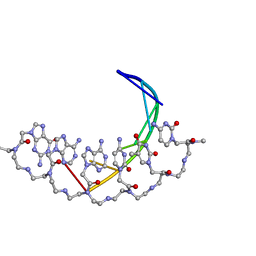 | |
8EBO
 
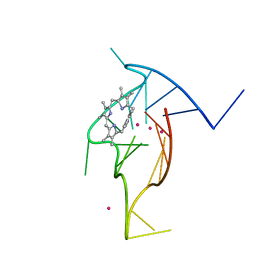 | | Homopurine parallel G-quadruplex from human chromosome 7 stabilized by K+ ions | | Descriptor: | COBALT (III) ION, DNA (5'-D(*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*AP*AP*AP*GP*GP*GP*GP*A)-3'), N-METHYLMESOPORPHYRIN, ... | | Authors: | Chen, E.V, Yatsunyk, L.A. | | Deposit date: | 2022-08-31 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Homopurine guanine-rich sequences in complex with N-methyl mesoporphyrin IX form parallel G-quadruplex dimers and display a unique symmetry tetrad.
Bioorg.Med.Chem., 77, 2022
|
|
8EDP
 
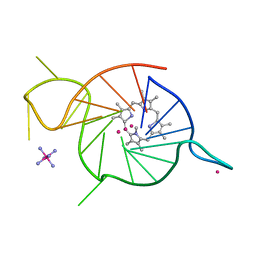 | | Crystal structure of a three-tetrad, parallel, and K+ stabilized homopurine G-quadruplex from human chromosome 7 | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*AP*GP*GP*GP*AP*AP*AP*AP*GP*GP*GP*GP*AP*GP*GP*GP*AP*AP*AP*AP*GP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, ... | | Authors: | Ye, M, Yatsunyk, L.A. | | Deposit date: | 2022-09-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Homopurine guanine-rich sequences in complex with N-methyl mesoporphyrin IX form parallel G-quadruplex dimers and display a unique symmetry tetrad.
Bioorg.Med.Chem., 77, 2022
|
|
1FHR
 
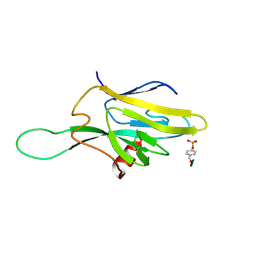 | | SOLUTION STRUCTURE OF THE FHA2 DOMAIN OF RAD53 COMPLEXED WITH A PHOSPHOTYROSYL PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Byeon, I.-J.L, Liao, H, Yongkiettrakul, S, Tsai, M.-D. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | II. Structure and specificity of the interaction between the FHA2 domain of Rad53 and phosphotyrosyl peptides.
J.Mol.Biol., 302, 2000
|
|
1US8
 
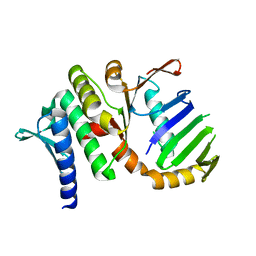 | | The Rad50 signature motif: essential to ATP binding and biological function | | Descriptor: | DNA DOUBLE-STRAND BREAK REPAIR RAD50 ATPASE | | Authors: | Moncalian, G, Lengsfeld, B, Bhaskara, V, Hopfner, K.P, Karcher, A, Alden, E, Tainer, J.A, Paull, T.T. | | Deposit date: | 2003-11-20 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Rad50 Signature Motif: Essential to ATP Binding and Biological Function
J.Mol.Biol., 335, 2004
|
|
2WCN
 
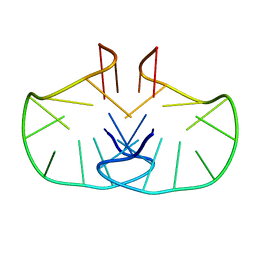 | | Solution structure of an LNA-modified quadruplex | | Descriptor: | DNA (5'-D(*DGP*LCG*DGP*LCG*DTP*DTP*DTP *DTP*DGP*LCG*DGP*LCG)-3') | | Authors: | Nielsen, J.T, Arar, K, Petersen, M. | | Deposit date: | 2009-03-12 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Locked Nucleic Acid Modified Quadruplex: Introducing the V4 Folding Topology.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2MA3
 
 | | NMR solution structure of the C-terminus of the minichromosome maintenance protein MCM from Methanothermobacter thermautotrophicus | | Descriptor: | DNA replication initiator (Cdc21/Cdc54) | | Authors: | Wiedemann, C, Ohlenschlager, O, Medagli, B, Onesti, S, Gorlach, M. | | Deposit date: | 2013-06-26 | | Release date: | 2014-12-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and regulatory role of the C-terminal winged helix domain of the archaeal minichromosome maintenance complex.
Nucleic Acids Res., 43, 2015
|
|
1YRM
 
 | |
3TAL
 
 | | Crystal structure of NurA with manganese | | Descriptor: | DNA double-strand break repair protein nurA, GLYCEROL, MANGANESE (II) ION | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
3TAI
 
 | | Crystal structure of NurA | | Descriptor: | DNA double-strand break repair protein nurA, GLYCEROL | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
2A7E
 
 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | DNA (5'-D(*CP*CP*CP*TP*AP*GP*GP*G)-3') | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7YQK
 
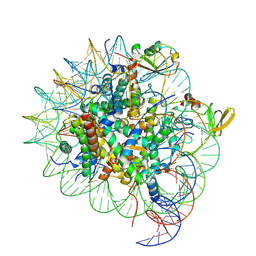 | | cryo-EM structure of gammaH2AXK15ub-H4K20me2 nucleosome bound to 53BP1 | | Descriptor: | DNA (145-MER), Histone H2AX, Histone H2B, ... | | Authors: | Ai, H.S, GuoChao, C, Qingyue, G, Ze-Bin, T, Zhiheng, D, Xin, L, Fan, Y, Ziyu, X, Jia-Bin, L, Changlin, T, Liu, L. | | Deposit date: | 2022-08-07 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Chemical Synthesis of Post-Translationally Modified H2AX Reveals Redundancy in Interplay between Histone Phosphorylation, Ubiquitination, and Methylation on the Binding of 53BP1 with Nucleosomes.
J.Am.Chem.Soc., 144, 2022
|
|
3IQS
 
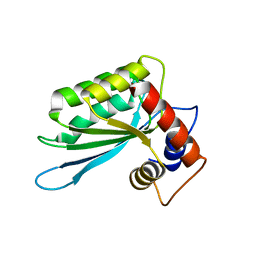 | | Crystal structure of the anti-viral APOBEC3G catalytic domain | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Holden, L.G, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
