1BJU
 
 | | BETA-TRYPSIN COMPLEXED WITH ACPU | | Descriptor: | 1-(4-AMIDINOPHENYL)-3-(4-CHLOROPHENYL)UREA, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Presnell, S, Patil, G, Mura, C, Jude, K, Conley, J, Kam, C, Bertrand, J, Powers, J, Williams, L. | | Deposit date: | 1998-06-29 | | Release date: | 1998-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxyanion-mediated inhibition of serine proteases.
Biochemistry, 37, 1998
|
|
4DED
 
 | | Aurora A in complex with YL1-038-21 | | Descriptor: | 1,2-ETHANEDIOL, 2-({2-[(4-carbamoylphenyl)amino]pyrimidin-4-yl}amino)benzamide, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Development of o-Chlorophenyl Substituted Pyrimidines as Exceptionally Potent Aurora Kinase Inhibitors.
J.Med.Chem., 55, 2012
|
|
3TUC
 
 | | Crystal structure of SYK kinase domain with 1-benzyl-N-(5-(6,7-dimethoxyquinolin-4-yloxy)pyridin-2-yl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Descriptor: | 1-benzyl-N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-2-oxo-1,2-dihydropyridine-3-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
3U71
 
 | | Crystal Structure Analysis of South African wild type HIV-1 Subtype C Protease | | Descriptor: | HIV-1 Protease | | Authors: | Naicker, P, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Dirr, H.W, Sayed, Y. | | Deposit date: | 2011-10-13 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure analysis of South African wild type HIV-1 subtype C apo protease
To be Published
|
|
3QPU
 
 | | PFKFB3 in complex with PPi | | Descriptor: | 1,2-ETHANEDIOL, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, PYROPHOSPHATE 2-, ... | | Authors: | Cavalier, M.C, Kim, S.G, Neau, D, Lee, Y.H. | | Deposit date: | 2011-02-14 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of the fructose-2,6-bisphosphatase reaction of PFKFB3: Transition state and the C-terminal function.
Proteins, 80, 2012
|
|
2QJG
 
 | | M. jannaschii ADH synthase complexed with F1,6P | | Descriptor: | 1,6-DI-O-PHOSPHONO-D-ALLITOL, Putative aldolase MJ0400 | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
3NTL
 
 | |
4HY5
 
 | | Crystal structure of cIAP1 BIR3 bound to T3256336 | | Descriptor: | (3S,7R,8aR)-2-{(2S)-2-(4,4-difluorocyclohexyl)-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]-7-ethoxyoctahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Snell, G.P. | | Deposit date: | 2012-11-13 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]pyrazine Scaffold as a Novel Proline Mimetic.
J.Med.Chem., 56, 2013
|
|
1J5M
 
 | | SOLUTION STRUCTURE OF THE SYNTHETIC 113CD_3 BETA_N DOMAIN OF LOBSTER METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Munoz, A, Forsterling, F.H, Shaw III, C.F, Petering, D.H. | | Deposit date: | 2002-05-16 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the (113)Cd(3)beta domains from Homarus americanus metallothionein-1: hydrogen bonding and solvent accessibility of sulfur atoms
J.Biol.Inorg.Chem., 7, 2002
|
|
4Q2B
 
 | | The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-D-glucanase, FORMIC ACID, ... | | Authors: | Tan, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440
To be Published
|
|
4LSV
 
 | | Crystal structure of broadly and potently neutralizing antibody 3BNC117 in complex with HIV-1 clade C C1086 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEAVY CHAIN OF ANTIBODY 3BNC117, IMIDAZOLE, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
3BOD
 
 | | Structure of mouse beta-neurexin 1 | | Descriptor: | CALCIUM ION, Neurexin-1-alpha | | Authors: | Koehnke, J, Jin, X, Shapiro, L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of beta-Neurexin 1 and beta-Neurexin 2 Ectodomains and Dynamics of Splice Insertion Sequence 4.
Structure, 16, 2008
|
|
4DEA
 
 | | Aurora A in complex with YL1-038-18 | | Descriptor: | 1,2-ETHANEDIOL, 4,4'-(pyrimidine-2,4-diyldiimino)dibenzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of o-Chlorophenyl Substituted Pyrimidines as Exceptionally Potent Aurora Kinase Inhibitors.
J.Med.Chem., 55, 2012
|
|
4Q4D
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMP-PNP and synthetic 3,5-(PP)2-IP4 (3,5-IP8) | | Descriptor: | (1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2014-04-14 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4DEB
 
 | | Aurora A in complex with RK2-17-01 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[3-(trifluoromethyl)phenyl]amino}pyrimidin-2-yl)amino]benzamide, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Development of o-Chlorophenyl Substituted Pyrimidines as Exceptionally Potent Aurora Kinase Inhibitors.
J.Med.Chem., 55, 2012
|
|
4HTN
 
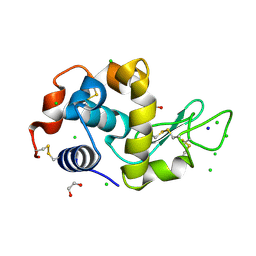 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 1.32 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1PI8
 
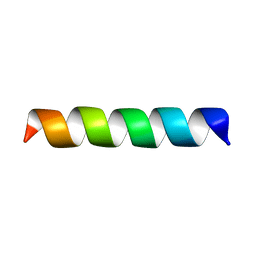 | | Structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1 | | Descriptor: | VPU protein | | Authors: | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2003-05-29 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Three-dimensional structure of the channel-forming trans-membrane domain of virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
4HY0
 
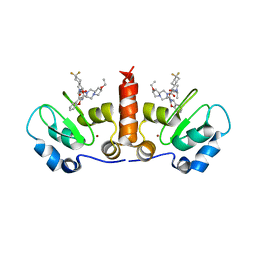 | | Crystal structure of XIAP BIR3 with T3256336 | | Descriptor: | (3S,7R,8aR)-2-{(2S)-2-(4,4-difluorocyclohexyl)-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]-7-ethoxyoctahydropyrrolo[1,2-a]pyrazine-3-carboxamide, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Snell, G.S, Dougan, D.R. | | Deposit date: | 2012-11-12 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]pyrazine Scaffold as a Novel Proline Mimetic.
J.Med.Chem., 56, 2013
|
|
1YY6
 
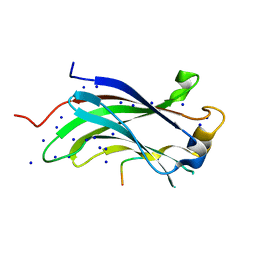 | | The Crystal Structure of the N-terminal domain of HAUSP/USP7 complexed with an EBNA1 peptide | | Descriptor: | Epstein-Barr nuclear antigen-1, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M, Shire, K, Nguyen, T, Zhang, R, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | Deposit date: | 2005-02-23 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
1PJE
 
 | | Structure of the channel-forming trans-membrane domain of Virus protein "u"(Vpu) from HIV-1 | | Descriptor: | VPU protein | | Authors: | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2003-06-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Three-dimensional structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
3ZHM
 
 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 in complex with the OL2 operator half-site | | Descriptor: | 5'-D(*AP*CP*GP*TP*GP*AP*AP*CP*TP*TP*GP*CP*AP*CP *TP*TP*GP*A)-3', 5'-D(*AP*GP*TP*TP*CP*AP*CP*GP*TP*TP*CP*AP*AP*GP *TP*GP*CP*A)-3', CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
3L8C
 
 | |
2QEZ
 
 | |
1CE0
 
 | |
3GET
 
 | |
