6V4A
 
 | |
3VTR
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 E328A complexed with TMG-chitotriomycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Chen, W, Liu, L, Shen, X, Yang, Q. | | Deposit date: | 2012-06-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into cellulolytic and chitinolytic enzymes revealing crucial residues of insect beta-N-acetyl-D-hexosaminidase
Plos One, 7, 2012
|
|
6ST6
 
 | | Crystal Structure of Domain Swapped Trp Repressor V58I Variant | | Descriptor: | ISOPROPYL ALCOHOL, Trp operon repressor | | Authors: | Sprenger, J, Lawson, C.L, Carey, J, Drouard, F, von Wachenfeldt, C, Schulz, A, Linse, S, Lo Leggio, L. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of Val58Ile tryptophan repressor in a domain-swapped array in the presence and absence of L-tryptophan.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6OZD
 
 | |
3VZJ
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) E172H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
6SAV
 
 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, CALCIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
4HSF
 
 | |
6P4G
 
 | | Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 1) | | Descriptor: | 18S rRNA, IAPV-IRES, RACK1, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
3FKY
 
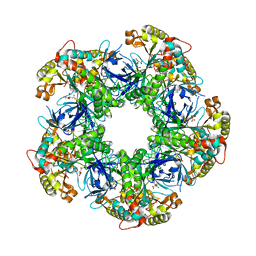 | | Crystal structure of the glutamine synthetase Gln1deltaN18 from the yeast Saccharomyces cerevisiae | | Descriptor: | CITRATE ANION, Glutamine synthetase | | Authors: | He, Y.X, Gui, L, Liu, Y.Z, Du, Y, Zhou, Y.Y, Li, P, Zhou, C.Z. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae glutamine synthetase Gln1 suggests a nanotube-like supramolecular assembly
Proteins, 76, 2009
|
|
6PMO
 
 | |
4DVH
 
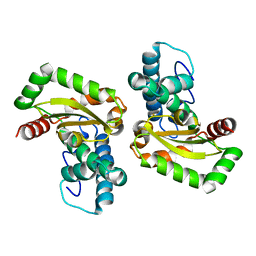 | | Crystal structure of Trypanosoma cruzi mitochondrial iron superoxide dismutase | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Larrieux, N, Buschiazzo, A. | | Deposit date: | 2012-02-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Molecular Basis of the Peroxynitrite-mediated Nitration and Inactivation of Trypanosoma cruzi Iron-Superoxide Dismutases (Fe-SODs) A and B: DISPARATE SUSCEPTIBILITIES DUE TO THE REPAIR OF TYR35 RADICAL BY CYS83 IN Fe-SODB THROUGH INTRAMOLECULAR ELECTRON TRANSFER.
J.Biol.Chem., 289, 2014
|
|
6SP7
 
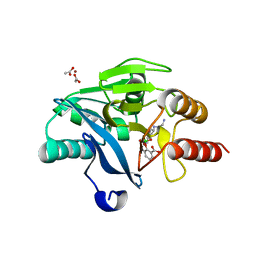 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with Taniborbactam (VNRX-5133) | | Descriptor: | (4~{R})-4-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, ACETATE ION, Metallo-beta-lactamase VIM-2, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Taniborbactam (VNRX-5133): A Broad-Spectrum Serine- and Metallo-beta-lactamase Inhibitor for Carbapenem-Resistant Bacterial Infections.
J.Med.Chem., 63, 2020
|
|
3EY8
 
 | | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, SULFATE ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1
To be Published
|
|
6V3Z
 
 | |
4I36
 
 | | Crystal Structure of the Bacillus stearothermophilus Phosphofructokinase Mutant D12A | | Descriptor: | 6-phosphofructokinase | | Authors: | Mosser, R, Reddy, M, Bruning, J.B, Sacchettini, J.C, Reinhart, G.D. | | Deposit date: | 2012-11-25 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redefining the Role of the Quaternary Shift in Bacillus stearothermophilus Phosphofructokinase.
Biochemistry, 52, 2013
|
|
3IC0
 
 | | Crystal Structure of liganded hemoglobin in complex with a potent antisickling agent, INN-298 | | Descriptor: | 2-[(2-methoxy-5-methylphenoxy)methyl]pyridine, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Safo, M.K, Musayev, F.N, Gandhi, A.K, Jorge, P. | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and in Vitro Chracterization of Pridyl Derivatives of Benzaldehydes: Highly Potent Antisickling Agents
To be Published
|
|
6S9S
 
 | | Dimerization domain of Xenopus laevis LDB1 in complex with darpin 10 | | Descriptor: | Darpin 10, LIM domain-binding protein 1 | | Authors: | Renko, M, Schaefer, J.V, Pluckthun, A, Bienz, M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-09 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rotational symmetry of the structured Chip/LDB-SSDP core module of the Wnt enhanceosome.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4I32
 
 | | Crystal structure of HCV NS3/4A D168V protease complexed with compound 4 | | Descriptor: | (2R,6S,7E,10E,13aR,14aR,16aS)-2-{[7-methoxy-8-methyl-2-(propan-2-yloxy)quinolin-4-yl]oxy}-N-[(1-methylcyclopropyl)sulfonyl]-6-{[(1-methyl-1H-pyrazol-3-yl)carbonyl]amino}-5,16-dioxo-1,2,3,6,9,12,13,13a,14,15,16,16a-dodecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamide, Genome polyprotein, HCV non-structural protein 4A, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-02 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | Molecular Mechanism by Which a Potent Hepatitis C Virus NS3-NS4A Protease Inhibitor Overcomes Emergence of Resistance.
J.Biol.Chem., 288, 2013
|
|
3F7O
 
 | | Crystal structure of Cuticle-Degrading Protease from Paecilomyces lilacinus (PL646) | | Descriptor: | (MSU)(ALA)(ALA)(PRO)(VAL), CALCIUM ION, Serine protease | | Authors: | Liang, L, Lou, Z, Meng, Z, Rao, Z, Zhang, K. | | Deposit date: | 2008-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
3IET
 
 | | Crystal Structure of 237mAb with antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Immunoglobulin heavy chain (IgG2a), Immunoglobulin light chain (IgG2a), ... | | Authors: | Brooks, C.L, Evans, S.V, Borisova, S.N. | | Deposit date: | 2009-07-23 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibody recognition of a unique tumor-specific glycopeptide antigen.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3F9N
 
 | | Crystal structure of chk1 kinase in complex with inhibitor 38 | | Descriptor: | 3-(3-chlorophenyl)-2-({(1S)-1-[(6S)-2,8-diazaspiro[5.5]undec-2-ylcarbonyl]pentyl}sulfanyl)quinazolin-4(3H)-one, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S, Ikuta, M. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of thioquinazolinones, allosteric Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6V33
 
 | | X-ray structure of a sugar N-formyltransferase from Pseudomonas congelans | | Descriptor: | 1,2-ETHANEDIOL, FOLIC ACID, dTDP-4-amino-4,6-dideoxyglucose, ... | | Authors: | Girardi, N.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Misannotations of the genes encoding sugar N-formyltransferases.
Protein Sci., 29, 2020
|
|
6P4X
 
 | |
6V7J
 
 | | The C2221 crystal form of canavalin at 173 K | | Descriptor: | BENZOIC ACID, CALCIUM ION, Canavalin, ... | | Authors: | McPherson, A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of benzoic acid and anions within the cupin domains of the vicilin protein canavalin from jack bean (Canavalia ensiformis): Crystal structures.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
6SAR
 
 | | E coli BepA/YfgC | | Descriptor: | Beta-barrel assembly-enhancing protease, SULFATE ION, ZINC ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2019-07-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Function Characterization of the Conserved Regulatory Mechanism of the Escherichia coli M48 Metalloprotease BepA.
J.Bacteriol., 203, 2020
|
|
