2MKM
 
 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3') | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-10 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
2KC9
 
 | |
4C41
 
 | |
2W7Z
 
 | | Structure of the pentapeptide repeat protein EfsQnr, a DNA gyrase inhibitor. Free amines modified by cyclic pentylation with glutaraldehyde. | | Descriptor: | CHLORIDE ION, PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Vetting, M.W, Hegde, S.S, Blanchard, J.S. | | Deposit date: | 2009-01-06 | | Release date: | 2009-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of a Pentapeptide-Repeat Protein by Reductive Cyclic Pentylation of Free Amines with Glutaraldehyde.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4BGB
 
 | |
4BGA
 
 | |
4CX8
 
 | | Monomeric pseudorabies virus protease pUL26N at 2.5 A resolution | | Descriptor: | PSEUDORABIES VIRUS PROTEASE | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-04 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease.
Plos Pathog., 11, 2015
|
|
2RAQ
 
 | | Crystal structure of the MTH889 protein from Methanothermobacter thermautotrophicus. Northeast Structural Genomics Consortium target TT205 | | Descriptor: | CALCIUM ION, Conserved protein MTH889 | | Authors: | Forouhar, F, Su, M, Xu, X, Seetharaman, J, Mao, L, Xiao, R, Ma, L.-C, Conover, K, Baran, M.C, Acton, T.B, Montelione, G.T, Arrowsmith, C.H, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-17 | | Release date: | 2007-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of the MTH889 protein from Methanothermobacter thermautotrophicus.
To be Published
|
|
2RFI
 
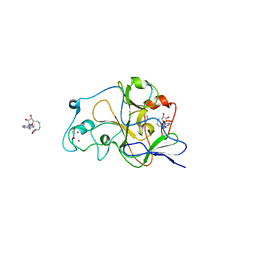 | | Crystal structure of catalytic domain of human euchromatic histone methyltransferase 1 in complex with SAH and dimethylated H3K9 peptide | | Descriptor: | Histone H3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Min, J, Wu, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
2VF2
 
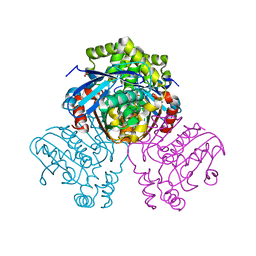 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2WGO
 
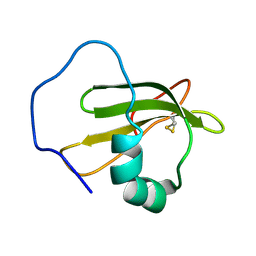 | | Structure of ranaspumin-2, a surfactant protein from the foam nests of a tropical frog | | Descriptor: | RANASPUMIN-2 | | Authors: | Mackenzie, C.D, Smith, B.O, Kennedy, M.W, Cooper, A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-06-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Ranaspumin-2: Structure and Function of a Surfactant Protein from the Foam Nests of a Tropical Frog.
Biophys.J., 96, 2009
|
|
2VVK
 
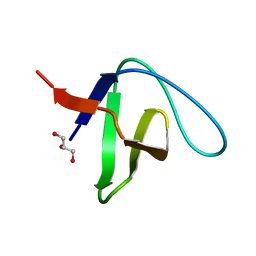 | | Grb2 SH3C (1) | | Descriptor: | GLYCEROL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2 | | Authors: | Harkiolaki, M, Tsirka, T, Feller, S.M. | | Deposit date: | 2008-06-09 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct Binding Modes of Two Epitopes in Gab2 that Interact with the Sh3C Domain of Grb2.
Structure, 17, 2009
|
|
1QRK
 
 | | HUMAN FACTOR XIII WITH STRONTIUM BOUND IN THE ION SITE | | Descriptor: | PROTEIN (COAGULATION FACTOR XIII), STRONTIUM ION | | Authors: | Fox, B.A, Yee, V.C, Pederson, L.C, Le Trong, I, Bishop, P.D, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 1999-06-14 | | Release date: | 1999-07-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of the calcium binding site and a novel ytterbium site in blood coagulation factor XIII by x-ray crystallography.
J.Biol.Chem., 274, 1999
|
|
1QA0
 
 | | BOVINE TRYPSIN 2-AMINOBENZIMIDAZOLE COMPLEX | | Descriptor: | 2H-BENZOIMIDAZOL-2-YLAMINE, CALCIUM ION, TRYPSIN | | Authors: | Whitlow, M. | | Deposit date: | 1999-04-09 | | Release date: | 2000-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2ZPK
 
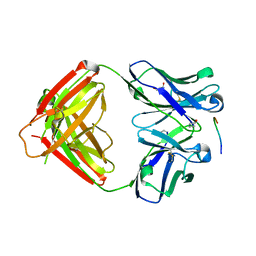 | | Crystal structure of P20.1 Fab fragment in complex with its antigen peptide | | Descriptor: | GLYCEROL, IgG1-lambda P20.1 Fab (heavy chain), IgG1-lambda P20.1 Fab (light chain), ... | | Authors: | Nogi, T, Sanagawa, T, Takagi, J. | | Deposit date: | 2008-07-16 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel affinity tag system using structurally defined antibody-tag interaction: application to single-step protein purification
Protein Sci., 17, 2008
|
|
1ZOM
 
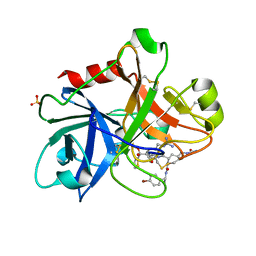 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in complex with a peptidomimetic Inhibitor | | Descriptor: | (S)-2-(3-((R)-1-(4-BROMOPHENYL)ETHYL)UREIDO)-N-((S)-1-((S)-5-GUANIDINO-1-OXO-1-(THIAZOL-2-YL)PENTAN-2-YLAMINO)-3-METHYL-1-OXOBUTAN-2-YL)-5-UREIDOPENTANAMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Lin, J, Deng, H, Jin, L, Pandey, P, Rynkiewicz, M, Bibbins, F, Cantin, S, Quinn, J, Magee, S, Gorga, J. | | Deposit date: | 2005-05-13 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design, synthesis, and biological evaluation of peptidomimetic inhibitors of factor XIa as novel anticoagulants.
J.Med.Chem., 49, 2006
|
|
1Z1V
 
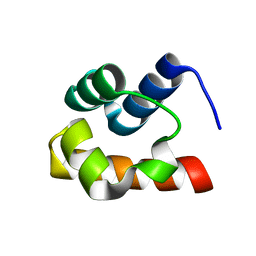 | | NMR structure of the Saccharomyces cerevisiae Ste50 SAM domain | | Descriptor: | STE50 protein | | Authors: | Kwan, J.J, Warner, N, Maini, J, Pawson, T, Donaldson, L.W. | | Deposit date: | 2005-03-06 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Saccharomyces cerevisiae Ste50 binds the MAPKKK Ste11 through a head-to-tail SAM domain interaction.
J.Mol.Biol., 356, 2006
|
|
1H1J
 
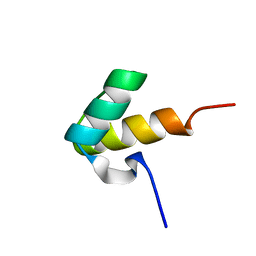 | |
1ZEO
 
 | | Crystal Structure of Human PPAR-gamma Ligand Binding Domain Complexed with an Alpha-Aryloxyphenylacetic Acid Agonist | | Descriptor: | (2S)-(4-ISOPROPYLPHENYL)[(2-METHYL-3-OXO-5,7-DIPROPYL-2,3-DIHYDRO-1,2-BENZISOXAZOL-6-YL)OXY]ACETATE, Peroxisome proliferator activated receptor gamma | | Authors: | Shi, G.Q, Dropinski, J.F, McKeever, B.M, Adams, A.D, MacNaul, K.L, Elbrecht, A, Berger, J.P, Zhou, G, Doebber, T.W. | | Deposit date: | 2005-04-19 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and Synthesis of alpha-Aryloxyphenylacetic Acid Derivatives: A Novel Class of PPAR alpha/gamma Dual Agonists with Potent Antihyperglycemic and Lipid Modulating Activity
J.Med.Chem., 48, 2005
|
|
1ZG8
 
 | | Crystal Structure of (R)-2-(3-{[amino(imino)methyl]amino}phenyl)-3-sulfanylpropanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | (2R)-2-(3-{[AMINO(IMINO)METHYL]AMINO}PHENYL)-3-SULFANYLPROPANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
2AIQ
 
 | |
2AQW
 
 | | Structure of putative orotidine-monophosphate-decarboxylase from Plasmodium yoelii (PY01515) | | Descriptor: | IODIDE ION, SULFATE ION, putative orotidine-monophosphate-decarboxylase | | Authors: | Dong, A, Vedadi, M, Wasney, G, Zhao, Y, Lew, J, Alam, Z, Melone, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-18 | | Release date: | 2005-08-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1RE3
 
 | | Crystal Structure of Fragment D of BbetaD398A Fibrinogen with the Peptide Ligand Gly-His-Arg-Pro-Amide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha/alpha-E chain, ... | | Authors: | Kostelansky, M.S, Betts, L, Gorkun, O.V, Lord, S.T. | | Deposit date: | 2003-11-06 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | BbetaGlu397 and BbetaAsp398 but not BbetaAsp432 are required for "B:b" interactions.
Biochemistry, 43, 2004
|
|
2BDD
 
 | | Crystal Structure of Holo-ACP-synthase from Plasmodium yoelii | | Descriptor: | ACP-synthase | | Authors: | Dong, A, Melone, M, Zhao, Y, Lew, J, Koeieradzki, I, Alam, Z, Wasney, G, Vedadi, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1RE4
 
 | | Crystal Structure of Fragment D of BbetaD398A Fibrinogen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha/alpha-E chain, ... | | Authors: | Kostelansky, M.S, Betts, L, Gorkun, O.V, Lord, S.T. | | Deposit date: | 2003-11-06 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | BbetaGlu397 and BbetaAsp398 but not BbetaAsp432 are required for "B:b" interactions.
Biochemistry, 43, 2004
|
|
