8ETI
 
 | | Fkbp39 associated 60S nascent ribosome State 1 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETH
 
 | | Ytm1 associated 60S nascent ribosome State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUP
 
 | | Ytm1 associated 60S nascent ribosome State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EV3
 
 | | Ytm1 associated 60S nascent ribosome (-Fkbp39) State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
3KKU
 
 | | Cruzain in complex with a non-covalent ligand | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, N-[2-(1H-benzimidazol-2-yl)ethyl]-2-(2-bromophenoxy)acetamide, ... | | Authors: | Ferreira, R.S, Eidam, O, Shoichet, B.K. | | Deposit date: | 2009-11-06 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Complementarity between a docking and a high-throughput screen in discovering new cruzain inhibitors.
J.Med.Chem., 53, 2010
|
|
3N8X
 
 | | Crystal Structure of Cyclooxygenase-1 in Complex with Nimesulide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ... | | Authors: | Lee, J.Y. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
3PH7
 
 | | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate | | Descriptor: | Farnesyl pyrophosphate synthase, GERANYLGERANYL DIPHOSPHATE | | Authors: | Wernimont, A.K, Dunford, J, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapiro, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites.
J.Biol.Chem., 286, 2011
|
|
3PHV
 
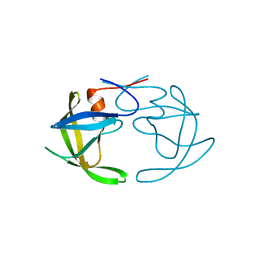 | | X-RAY ANALYSIS OF HIV-1 PROTEINASE AT 2.7 ANGSTROMS RESOLUTION CONFIRMS STRUCTURAL HOMOLOGY AMONG RETROVIRAL ENZYMES | | Descriptor: | UNLIGANDED HIV-1 PROTEASE | | Authors: | Lapatto, R, Blundell, T.L, Hemmings, A, Wilderspin, A, Wood, S.P, Danley, D.E, Geoghegan, K.F, Hawrylik, S.J, Hobart, P.M. | | Deposit date: | 1991-11-04 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes.
Nature, 342, 1989
|
|
4F3T
 
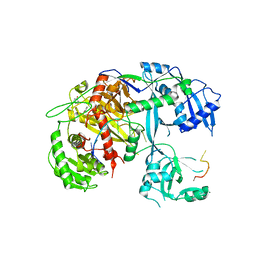 | | Human Argonaute-2 - miR-20a complex | | Descriptor: | PHENOL, Protein argonaute-2, RNA (5'-R(P*UP*AP*AP*AP*GP*UP*GP*CP*UP*UP*AP*UP*AP*GP*UP*G*CP*AP*GP*G)-3') | | Authors: | Elkayam, E, Kuhn, C.-D, Tocilj, A, Joshua-Tor, L. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of Human Argonaute-2 in Complex with miR-20a.
Cell(Cambridge,Mass.), 150, 2012
|
|
6WTH
 
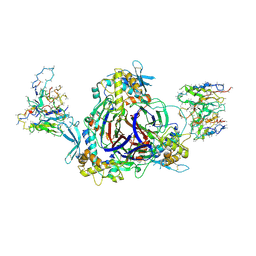 | | Full-length human ENaC ECD | | Descriptor: | 10D4 Fab, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Posert, R, Baconguis, I, Noreng, S, Bharadwaj, A, Houser, A. | | Deposit date: | 2020-05-02 | | Release date: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Molecular principles of assembly, activation, and inhibition in epithelial sodium channel.
Elife, 9, 2020
|
|
6WM3
 
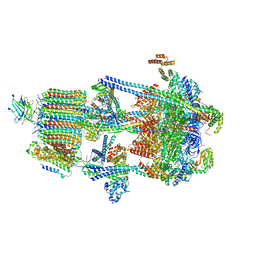 | | Human V-ATPase in state 2 with SidK and ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Renin receptor, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
3RC9
 
 | |
1EZU
 
 | | ECOTIN Y69F, D70P BOUND TO D102N TRYPSIN | | Descriptor: | CALCIUM ION, ECOTIN, TRYPSIN II, ... | | Authors: | Gillmor, S.A, Takeuchi, T, Yang, S.Q, Craik, C.S, Fletterick, R.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-06-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases.
J.Mol.Biol., 299, 2000
|
|
1EZS
 
 | | CRYSTAL STRUCTURE OF ECOTIN MUTANT M84R, W67A, G68A, Y69A, D70A BOUND TO RAT ANIONIC TRYPSIN II | | Descriptor: | CALCIUM ION, ECOTIN, TRYPSIN II, ... | | Authors: | Gillmor, S.A, Takeuchi, T, Yang, S.Q, Craik, C.S, Fletterick, R.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-06-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases.
J.Mol.Biol., 299, 2000
|
|
7S4G
 
 | | Fab fragment bound to the Cter peptide of Ly6G6D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Lymphocyte antigen 6 complex locus protein G6d, ... | | Authors: | Rouge, L, Lupardus, P. | | Deposit date: | 2021-09-08 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Anti-LY6G6D/CD3 T-Cell-Dependent Bispecific Antibody for the Treatment of Colorectal Cancer.
Mol.Cancer Ther., 21, 2022
|
|
8I9P
 
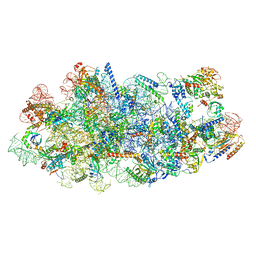 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Mak16 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, 60S ribosomal protein L16-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
7SH4
 
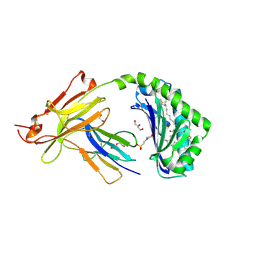 | | CD1a-phosphatidylglycerol binary structure | | Descriptor: | (21R,24R,27S)-24,27,28-trihydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaoctacosan-21-yl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wegrecki, M, Rossjohn, J. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylococcal phosphatidylglycerol antigens activate human T cells via CD1a.
Nat.Immunol., 24, 2023
|
|
2R63
 
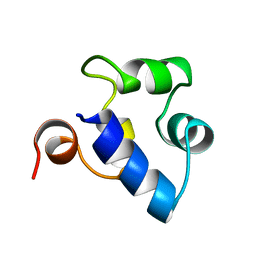 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
3J7P
 
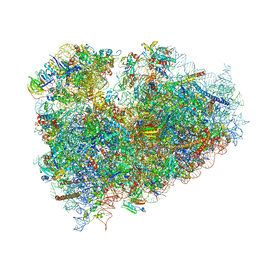 | | Structure of the 80S mammalian ribosome bound to eEF2 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-03-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7SUT
 
 | | Light harvesting phycobiliprotein HaPE645 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | (15,16)-DIHYDROBILIVERDIN (SINGLY LINKED), 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-18 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
3ZMB
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound fragment SPB02696. | | Descriptor: | 3-(2-oxo-1,3-benzoxazol-3(2H)-yl)propanoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2013-02-07 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8FKZ
 
 | |
