3MEE
 
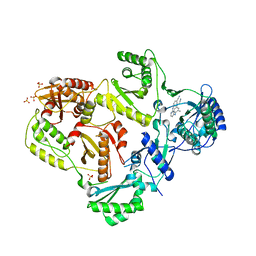 | | HIV-1 Reverse Transcriptase in Complex with TMC278 | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, SULFATE ION, p51 Reverse transcriptase, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
6ZNL
 
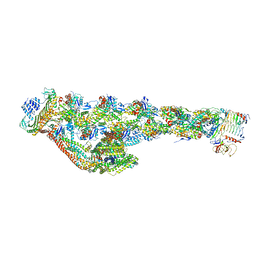 | | Cryo-EM structure of the dynactin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Lacey, S.E, Carter, A.P. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
8QKW
 
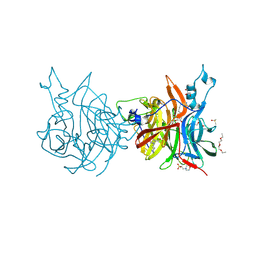 | | Crystal structure of Levansucrase from Pseudomonas syringae in complex with a tetravalent iminosugar | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-(hydroxymethyl)-1-[6-[4-[[3-[[3-[6-[(2~{S},3~{R},4~{S})-2-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidin-1-yl]hexyl]-1,2,3-triazol-4-yl]methoxy]-2,2-bis[[1-[6-[2-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidin-1-yl]hexyl]-1,2,3-triazol-4-yl]methoxymethyl]propoxy]methyl]-1,2,3-triazol-1-yl]hexyl]pyrrolidine-3,4-diol, DIMETHYL SULFOXIDE, ... | | Authors: | Ferraroni, M, Canovai, A. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Levansucrase from Pseudomonas syringae in complex with a tetravalent iminosugar
To Be Published
|
|
5GAE
 
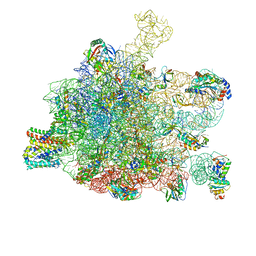 | | RNC in complex with a translocating SecYEG | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
6ZR2
 
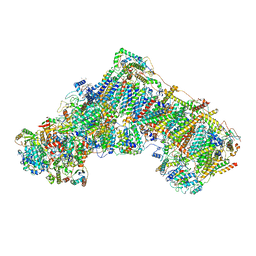 | | Cryo-EM structure of respiratory complex I in the active state from Mus musculus at 3.1 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | Deposit date: | 2020-07-10 | | Release date: | 2020-10-21 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
7SBT
 
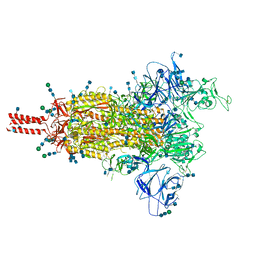 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Gamma variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
3M78
 
 | |
4G0L
 
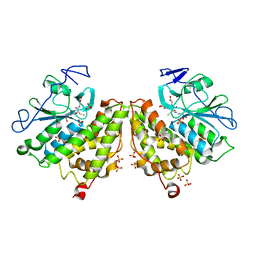 | | Glutathionyl-hydroquinone Reductase, YqjG, of E.coli complexed with GSH | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
8QEW
 
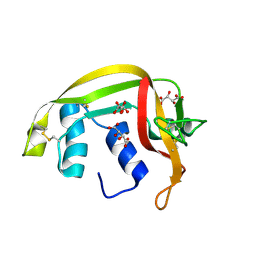 | |
4G0V
 
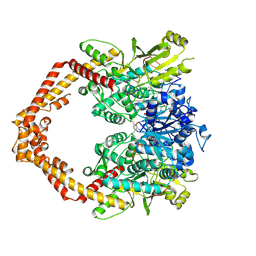 | | Human topoisomerase iibeta in complex with DNA and mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Wu, C.C, Li, T.K, Li, Y.C, Chan, N.L. | | Deposit date: | 2012-07-10 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs
Nucleic Acids Res., 41, 2013
|
|
4G1N
 
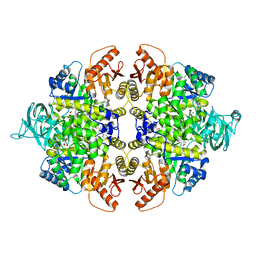 | | PKM2 in complex with an activator | | Descriptor: | MAGNESIUM ION, N-(4-{[4-(pyrazin-2-yl)piperazin-1-yl]carbonyl}phenyl)quinoline-8-sulfonamide, OXALATE ION, ... | | Authors: | Kung, C, Hixon, J, Dang, L, DeLaBarre, B, Qian, K.C. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small Molecule Activation of PKM2 in Cancer Cells Induces Serine Auxotrophy.
Chem.Biol., 19, 2012
|
|
8QSR
 
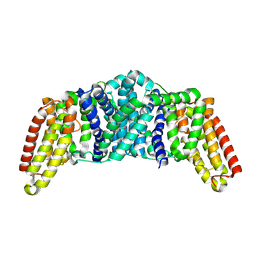 | |
8Q0G
 
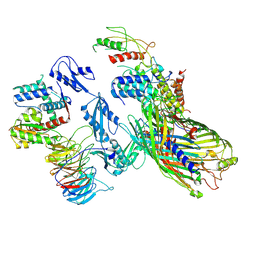 | | Release Complex: BAM bound EspP and Compact SurA | | Descriptor: | Chaperone SurA, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Fenn, K.L, Ranson, N.A. | | Deposit date: | 2023-07-28 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Outer membrane protein assembly mediated by BAM-SurA complexes.
Nat Commun, 15, 2024
|
|
3ZR1
 
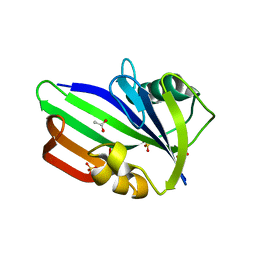 | | Crystal structure of human MTH1 | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, SULFATE ION | | Authors: | Svensson, L.M, Jemth, A, Desroses, M, Loseva, O, Helleday, T, Hogbom, M, Stenmark, P. | | Deposit date: | 2011-06-13 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Mth1 and the 8-Oxo-Dgmp Product Complex.
FEBS Lett., 585, 2011
|
|
7N78
 
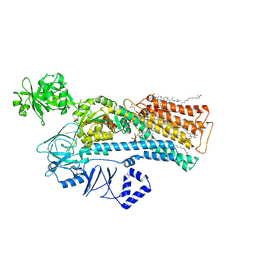 | | Cryo-EM structure of ATP13A2 in the E2-Pi state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sim, S.I, Park, E. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of polyamine transport by human ATP13A2 (PARK9).
Mol.Cell, 81, 2021
|
|
5GKN
 
 | |
4G4G
 
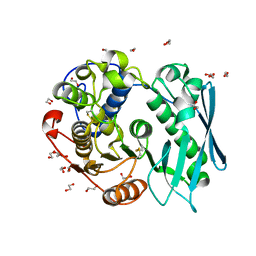 | | Crystal structure of recombinant glucuronoyl esterase from Sporotrichum thermophile determined at 1.55 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-glucuronoyl methylesterase, GLYCEROL | | Authors: | Charavgi, M.D, Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2012-07-16 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6PD9
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 60 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-(1H-pyrazol-1-yl)benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
3ZR0
 
 | | Crystal structure of human MTH1 in complex with 8-oxo-dGMP | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION | | Authors: | Svensson, L.M, Jemth, A, Desroses, M, Loseva, O, Helleday, T, Hogbom, M, Stenmark, P. | | Deposit date: | 2011-06-13 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Mth1 and the 8-Oxo-Dgmp Product Complex.
FEBS Lett., 585, 2011
|
|
6ZIY
 
 | | Respiratory complex I from Thermus thermophilus, NADH dataset, major state | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6PDF
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 55 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-(2H-1,2,3-triazol-2-yl)benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
3ZSK
 
 | | Crystal structure of Human Galectin-3 CRD with glycerol bound at 0.90 angstrom resolution | | Descriptor: | GALECTIN-3, GLYCEROL | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZR6
 
 | | STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE IN COMPLEX WITH GALACTOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Deane, J.E, Graham, S.C, Kim, N.N, Stein, P.E, Mcnair, R, Cachon-Gonzalez, M.B, Cox, T.M, Read, R.J. | | Deposit date: | 2011-06-14 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Insights Into Krabbe Disease from Structures of Galactocerebrosidase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6ZK0
 
 | | 1.47A human IMPase with ebselen | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Inositol monophosphatase 1, ... | | Authors: | Bax, B.D, Fenn, G.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallization and structure of ebselen bound to Cys141 of human inositol monophosphatase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7S5X
 
 | | Human KATP channel in open conformation, focused on Kir and one SUR, position 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2021-09-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular structure of an open human K ATP channel.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
