7DTE
 
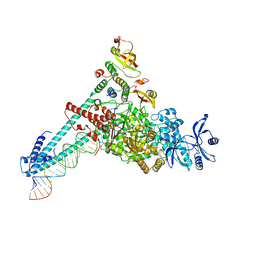 | | SARS-CoV-2 RdRP catalytic complex with T33-1 RNA | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (33-MER), ... | | Authors: | Wang, Q, Gong, P. | | Deposit date: | 2021-01-04 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex.
Cell Rep, 37, 2021
|
|
5I3P
 
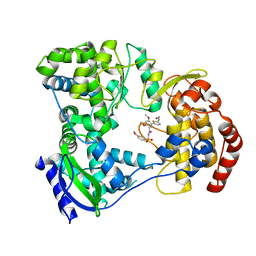 | | DENGUE SEROTYPE 3 RNA-DEPENDENT RNA POLYMERASE BOUND TO COMPOUND 27 | | Descriptor: | 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-2,4-dimethoxy-N-[(3-methoxyphenyl)sulfonyl]benzamide, Genome polyprotein, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-02-10 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
1YSV
 
 | |
5I3Q
 
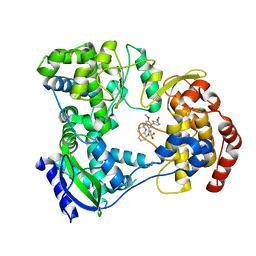 | | DENGUE SEROTYPE 3 RNA-DEPENDENT RNA POLYMERASE BOUND TO COMPOUND 29 | | Descriptor: | 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-4-methoxy-2-methyl-N-[(quinolin-8-yl)sulfonyl]benzamide, Genome polyprotein, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-02-10 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
8PSN
 
 | |
8PSO
 
 | | Tilapia Lake Virus polymerase in vRNA initiation state (core only) | | Descriptor: | 5' vRNA end - vRNA loop (40-mer), DNA (5'-D(*(CTP))-3'), MAGNESIUM ION, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2023-07-13 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural and functional analysis of the minimal orthomyxovirus-like polymerase of Tilapia Lake Virus from the highly diverged Amnoonviridae family.
Nat Commun, 14, 2023
|
|
8PSS
 
 | |
8PT2
 
 | |
8PT7
 
 | |
8PTJ
 
 | |
7EJU
 
 | | Junin virus(JUNV) RNA polymerase L complexed with Z protein | | Descriptor: | MAGNESIUM ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Chen, Y. | | Deposit date: | 2021-04-02 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for recognition and regulation of arenavirus polymerase L by Z protein.
Nat Commun, 12, 2021
|
|
8PSQ
 
 | |
8PSU
 
 | |
8PTH
 
 | |
8PSX
 
 | |
8PT6
 
 | |
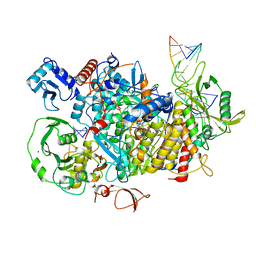
8PSZ
 
 | |
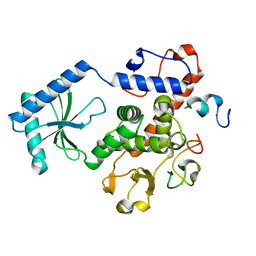
3NYB
 
 | |
2WK4
 
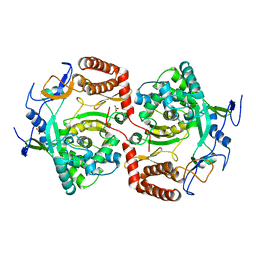 | | Dimeric structure of D347G D348G mutant of the sapporovirus RNA dependent RNA polymerase | | Descriptor: | GLYCEROL, PROTEASE-POLYMERASE P70 | | Authors: | Fullerton, S.W.B, Robel, I, Schuldt, L, Gebhardt, J, Tucker, P.A, Rohayem, J. | | Deposit date: | 2009-06-05 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Dimeric Structure of D347G D348G Mutant of the Sapporovirus Sapporovirus RNA Dependent RNA Polymerase
To be Published
|
|
2R7W
 
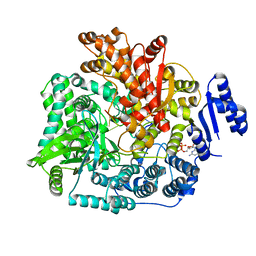 | | Crystal Structure of Rotavirus SA11 VP1/RNA (UGUGACC)/mRNA 5'-CAP (m7GpppG) complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA (5'-R(*UP*GP*UP*GP*AP*CP*C)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
8QZ8
 
 | |
5U04
 
 | | Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase | | Descriptor: | PHOSPHATE ION, ZINC ION, Zika Virus NS5 RdRp | | Authors: | Godoy, A.S, Lima, G.M.A, Oliveira, K.I.Z, Torres, N.U, Maluf, F.V, Guido, R.V.C, Oliva, G. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase.
Nat Commun, 8, 2017
|
|
5TRH
 
 | |
5TRI
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 3-[(4-chlorophenyl)methoxy]-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)benzoic acid | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 3-[(4-chlorophenyl)methoxy]-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)benzoic acid, NS5B RNA-DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and initial optimization of alkoxyanthranilic acid derivatives as inhibitors of HCV NS5B polymerase.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4FLB
 
 | | CID of human RPRD2 | | Descriptor: | PRASEODYMIUM ION, Regulation of nuclear pre-mRNA domain-containing protein 2, SULFATE ION, ... | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
