6FHJ
 
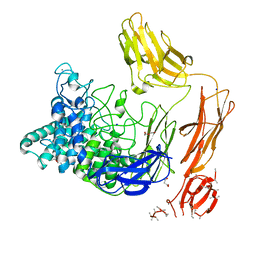 | | Structural dynamics and catalytic properties of a multi-modular xanthanase, native. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
7N1P
 
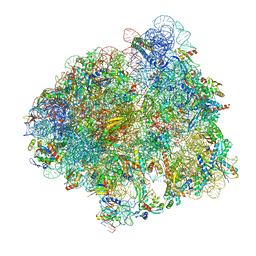 | | Elongating 70S ribosome complex in a classical pre-translocation (PRE-C) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, S.K, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
6OGI
 
 | | 70S termination complex with RF2 bound to the UAG codon. Rotated ribosome conformation (Structure V) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6OI2
 
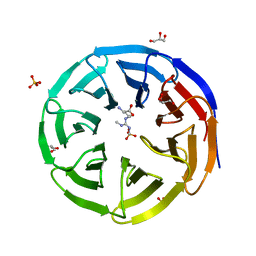 | | Crystal structure of human WDR5 in complex with symmetric dimethyl-L-arginine | | Descriptor: | GLYCEROL, N3, N4-DIMETHYLARGININE, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
4ER3
 
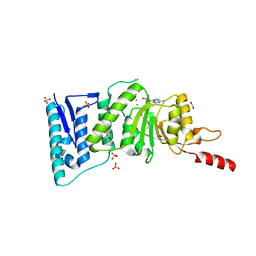 | | Crystal Structure of Human DOT1L in complex with inhibitor EPZ004777 | | Descriptor: | 1,2-ETHANEDIOL, 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
5FOW
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH WHTA TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION, WHTA PEPTIDE | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
6SVY
 
 | | Crystal structure of Neprilysin in complex with Sampatrilat-ASP. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, Sampatrilat-Asp, ... | | Authors: | Cozier, G.E, Acharya, K.R, Sharma, U. | | Deposit date: | 2019-09-19 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for Omapatrilat and Sampatrilat Binding to Neprilysin-Implications for Dual Inhibitor Design with Angiotensin-Converting Enzyme.
J.Med.Chem., 63, 2020
|
|
7O0T
 
 | | Crystal structure of Chloroflexus aggregans ene-reductase CaOYE holoenzyme | | Descriptor: | FLAVIN MONONUCLEOTIDE, MALONATE ION, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Robescu, M.S, Niero, M, Loprete, G, Bergantino, E, Cendron, L. | | Deposit date: | 2021-03-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A New Thermophilic Ene-Reductase from the Filamentous Anoxygenic Phototrophic Bacterium Chloroflexus aggregans .
Microorganisms, 9, 2021
|
|
7NQL
 
 | | 55S mammalian mitochondrial ribosome with ICT1 and P site tRNAMet | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-01 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7NQH
 
 | | 55S mammalian mitochondrial ribosome with mtRF1a and P-site tRNAMet | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-01 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
5D7I
 
 | | Structure of human MR1-Ac-6-FP in complex with human MAIT M33.64 TCR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, M33.64 TCR Alpha Chain, ... | | Authors: | Keller, A.N, Woolley, R.E, Rossjohn, J. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
7XC2
 
 | | Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 and the effector AvrSr35 of the wheat stem rust pathogen | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Avirulence factor, CNL9 | | Authors: | Alexander, F, Li, E.T, Aaron, L, Deng, Y.N, Sun, Y, Chai, J.J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A wheat resistosome defines common principles of immune receptor channels.
Nature, 610, 2022
|
|
5FOV
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHTG TETRAPEPTIDE | | Descriptor: | DNA repair and recombination protein RadA, FHTG PEPTIDE, GLYCEROL, ... | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
6Q9W
 
 | | X-ray structure of compound 15 bound to HdmX: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | (4~{S})-4-(4-chlorophenyl)-5-[(1~{S})-1-(3-chlorophenyl)ethyl]-2-(2,4-dimethoxypyrimidin-5-yl)-3-propan-2-yl-4~{H}-pyrrolo[3,4-d]imidazol-6-one, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
6G9I
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with clomipramine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(3-CHLORO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, Envelope glycoprotein,Envelope glycoprotein, ... | | Authors: | Zhao, Y, Ren, J, Fry, E.E, Xiao, J, Townsend, A.R, Stuart, D.I. | | Deposit date: | 2018-04-10 | | Release date: | 2018-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
J. Med. Chem., 61, 2018
|
|
5ER4
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC0025 | | Descriptor: | 6-methyl-5,6,6~{a},7-tetrahydro-4~{H}-dibenzo[de,g]quinoline-10,11-diol, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Melville, Z.E, Aligholizadeh, E, Fang, L, Alasady, M.J, Pierce, A.D, Wilder, P.T, MacKerell Jr, A.D, Weber, D.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Novel protein-inhibitor interactions in site 3 of Ca(2+)-bound S100B as discovered by X-ray crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7NSH
 
 | | 39S mammalian mitochondrial large ribosomal subunit with mtRRF (post) and mtEFG2 | | Descriptor: | 16S rRNA, 39S ribosomal protein L48, mitochondrial, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-07 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
5NFE
 
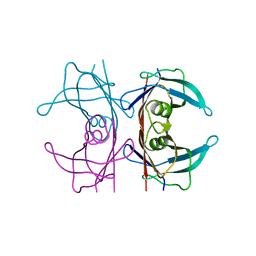 | | Neutron structure of human transthyretin (TTR) T119M mutant at room temperature to 1.85A resolution | | Descriptor: | Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Ostermann, A, Cooper, J.B, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2017-03-14 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.853 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
4WEN
 
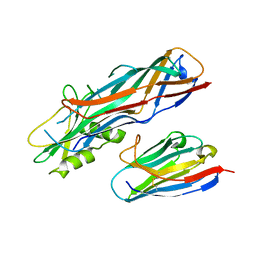 | | Co-complex structure of the F4 fimbrial adhesin FaeG variant ac with llama single domain antibody V2 | | Descriptor: | Anti-F4+ETEC bacteria VHH variable region, K88 fimbrial protein AC | | Authors: | Moonens, K, Van den Broeck, I, Pardon, E, De Kerpel, M, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic Escherichia coli by llama single domain antibodies.
Vet. Res., 46, 2015
|
|
8B0H
 
 | | 2C9, C5b9-CD59 cryoEM structure | | Descriptor: | CD59 glycoprotein, Complement C5, Complement component C6, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
1Y1K
 
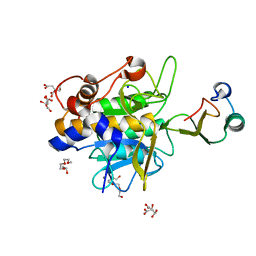 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 T58A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-18 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
6PXM
 
 | | Horse spleen apoferritin light chain | | Descriptor: | Ferritin light chain | | Authors: | Kopylov, M, Kelley, K, Yen, L.Y, Rice, W.J, Eng, E.T, Carragher, B, Potter, C.S. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Horse spleen apoferritin light chain structure at 2.1 Angstrom resolution
To Be Published
|
|
6G4F
 
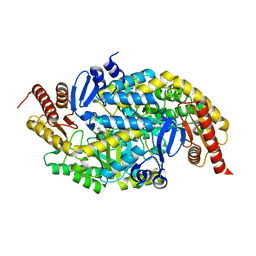 | |
1Y3F
 
 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 F69A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-24 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
6NAG
 
 | |
