7OR2
 
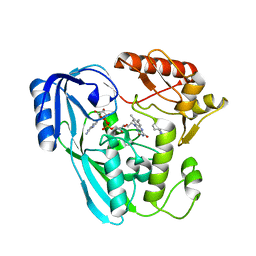 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 4) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7U08
 
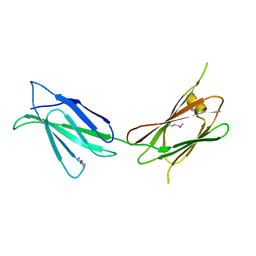 | |
7OSQ
 
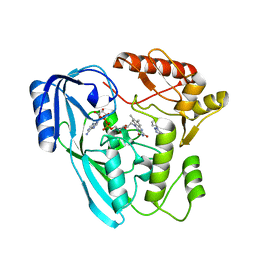 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 5-methyl-1-phenyl-1,2,3-triazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Mayol-Llinas, J, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
8TYY
 
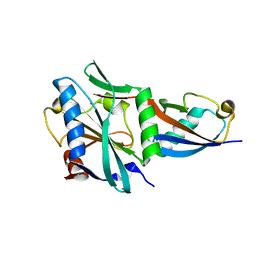 | |
7TZL
 
 | | The DH dehydratase domain of AlnB | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Swain, K, Blackson, W, Wang, B, Zhao, H, Nannenga, B.L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The programming of alpha,beta-polyene biosynthesis by a bacterial iterative type I polyketide synthase
To Be Published
|
|
5TKH
 
 | | Neurospora crassa polysaccharide monooxygenase 2 ascorbate treated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Lytic polysaccharide monooxygenase, ... | | Authors: | O'Dell, W.B, Meilleur, F. | | Deposit date: | 2016-10-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Oxygen Activation at the Active Site of a Fungal Lytic Polysaccharide Monooxygenase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5FUW
 
 | | catalytic domain of Thymidine kinase from Trypanosoma brucei with dTMP or dThd | | Descriptor: | PHOSPHATE ION, THYMDINE KINASE, THYMIDINE, ... | | Authors: | Timm, J, Valente, M, Castillo-Acosta, V, Balzarini, T, Nettleship, J.E, Rada, H, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2016-01-31 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cell Cycle Regulation and Novel Structural Features of Thymidine Kinase, an Essential Enzyme in Trypanosoma Brucei.
Mol.Microbiol., 102, 2016
|
|
8TCT
 
 | | Structure of 3K-GlcH bound Bacteroides thetaiotaomicron 3-Keto-beta-glucopyranoside-1,2-Lyase BT1 | | Descriptor: | 1,5-anhydro-D-ribo-hex-3-ulose, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
6MC2
 
 | |
7OOL
 
 | | Crystal structure of a Candidatus photodesmus katoptron thioredoxin chimera containing an ancestral loop | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gavira, J.A, Ibarra-Molero, B, Gamiz-Arco, G, Risso, V, Sanchez-Ruiz, J.M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining Ancestral Reconstruction with Folding-Landscape Simulations to Engineer Heterologous Protein Expression.
J.Mol.Biol., 433, 2021
|
|
7MPM
 
 | | Bartonella henselae NrnC bound to pAA | | Descriptor: | 5'-phosphorylated AA, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPN
 
 | | Bartonella henselae NrnC bound to pGC | | Descriptor: | 5'-phosphorylated GC, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
5TCW
 
 | |
5FLD
 
 | |
8TU3
 
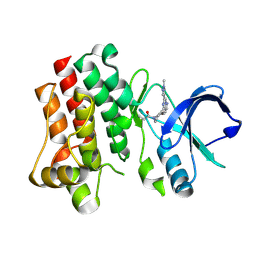 | | Bruton's tyrosine kinase in complex with covalent inhibitor compound 10 | | Descriptor: | 1-[(4R)-4-{[(6P,8S)-6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]oxy}azepan-1-yl]propan-1-one, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB129, a Covalent, Selective, and Brain-Penetrant BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 67, 2024
|
|
7P8Z
 
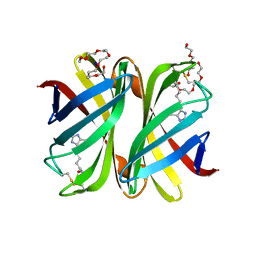 | | Short wilavidin biotin complex | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, BIOTIN, ... | | Authors: | Avraham, O, Livnah, O. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Wilavidin - a novel member of the avidin family that forms unique biotin-binding hexamers.
Febs J., 289, 2022
|
|
7MPQ
 
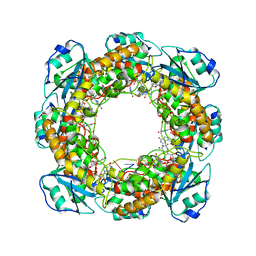 | |
7U0W
 
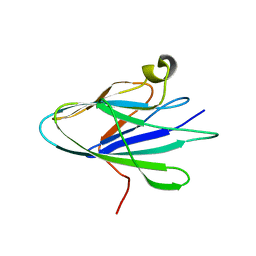 | |
5FTA
 
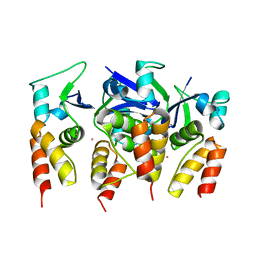 | | Crystal structure of the N-terminal BTB domain of human KCTD10 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 3, MERCURY (II) ION | | Authors: | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Tallant, C, Newman, J.A, Kopec, J, Fitzpatrick, F, Talon, R, Collins, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
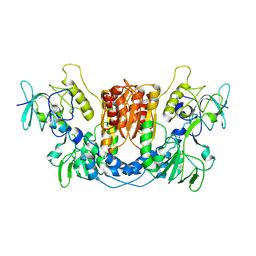
8QCK
 
 | |
8TZ0
 
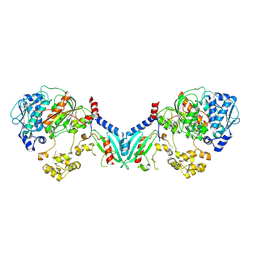 | | Structure of a bacterial E1-E2-Ubl complex (form 1) | | Descriptor: | E1(BilD), E2(BilB), Ubl(BilA), ... | | Authors: | Ye, Q, Chambers, L.R, Corbett, K.D. | | Deposit date: | 2023-08-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | A eukaryotic-like ubiquitination system in bacterial antiviral defence.
Nature, 631, 2024
|
|
5BYL
 
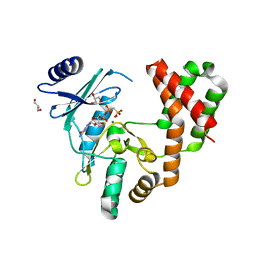 | |
7U0V
 
 | |
7U01
 
 | | Structure of CD148 fibronectin type III domain 2 | | Descriptor: | Receptor-type tyrosine-protein phosphatase eta | | Authors: | Zhou, D, Zhu, J. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.297079 Å) | | Cite: | Structure of CD148 fibronectin type III domain 1 and 2
To Be Published
|
|
8Q9P
 
 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC5 deacetylase binding motif | | Descriptor: | HDAC5 (histone deacetylase 5) binding motif peptide: TRP-GLY-SER-GLY-GLU-VAL-LYS-LEU-ARG-LEU-GLN-GLU-PHE-LEU-LEU-SER-LYS-SER, MADS box dsDNA fw: AACTATTTATAAGA, MADS box dsNA rev:TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
