2BU3
 
 | | Acyl-enzyme intermediate between Alr0975 and glutathione at pH 3.4 | | Descriptor: | ALR0975 PROTEIN, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vivares, D, Arnoux, P, Pignol, D. | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Papain-Like Enzyme at Work: Native and Acyl- Enzyme Intermediate Structures in Phytochelatin Synthesis.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1OUV
 
 | | Helicobacter cysteine rich protein C (HcpC) | | Descriptor: | conserved hypothetical secreted protein | | Authors: | Mittl, P.R, Luethy, L. | | Deposit date: | 2003-03-25 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Helicobacter Cysteine-rich Protein C at 2.0A Resolution: Similar Peptide-binding Sites in TPR and SEL1-like Repeat Proteins
J.Mol.Biol., 340, 2004
|
|
2NQD
 
 | | Crystal structure of cysteine protease inhibitor, chagasin, in complex with human cathepsin L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Cathepsin L, ... | | Authors: | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | Deposit date: | 2006-10-31 | | Release date: | 2007-07-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
9MEX
 
 | | Structure of phosphocysteine intermediate of human PRL1 phosphatase | | Descriptor: | Protein tyrosine phosphatase type IVA 1, SULFATE ION | | Authors: | Mahbub, L, Kozlov, G, Knorn, C, Gehring, K. | | Deposit date: | 2024-12-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the phosphocysteine intermediate of the phosphatase of regenerating liver PTP4A1.
J.Biol.Chem., 301, 2025
|
|
5K22
 
 | |
6UK3
 
 | |
6PO4
 
 | | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII. | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII.
To Be Published
|
|
6WB8
 
 | | Cryo-EM structure of PKD2 C331S disease variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Cao, E, Wang, J, Decaen, P.G. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Molecular dysregulation of ciliary polycystin-2 channels caused by variants in the TOP domain.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4BZP
 
 | | Structure of the Mycobacterium tuberculosis APS kinase CysC in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BIFUNCTIONAL ENZYME CYSN/CYSC | | Authors: | Poyraz, O, Lohkamp, B, Schnell, R, Schneider, G. | | Deposit date: | 2013-07-29 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structures of the Kinase Domain of the Sulfate-Activating Complex in Mycobacterium Tuberculosis.
Plos One, 10, 2015
|
|
5JCU
 
 | |
2H7W
 
 | |
6PI9
 
 | | Crystal structure of 16S rRNA methyltransferase RmtF in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 16S rRNA (guanine(1405)-N(7))-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Stogios, P.J, Kim, Y, Evdokimova, E, Di Leo, R, Semper, C, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 16S rRNA methylase RmtF in complex with S-Adenosyl-L-homocysteine
To be Published
|
|
9HKW
 
 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 3 open and 1 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 3 open and 1 closed subunits
To Be Published
|
|
9HKY
 
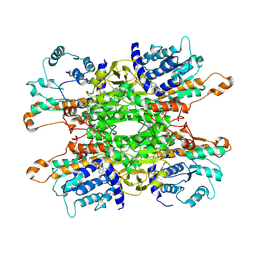 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 open and 2 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM struture of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 open and 2 closed subunits
To Be Published
|
|
9HKZ
 
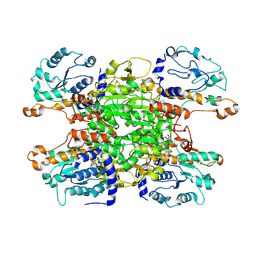 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 wide open, 1 open and 1 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 wide open, 1 open and 1 closed subunits
To Be Published
|
|
9HKX
 
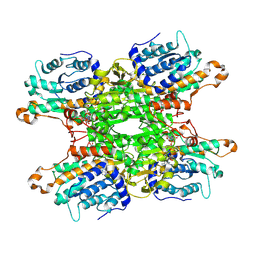 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 1 open and 3 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 1 open and 3 closed subunits
To Be Published
|
|
4GVO
 
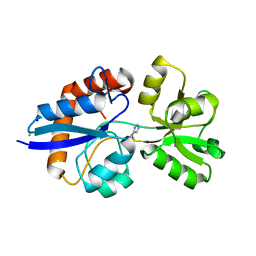 | | Putative L-Cystine ABC transporter from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, HISTIDINE, Lmo2349 protein, ... | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Putative L-Cystine ABC transporter from Listeria monocytogenes
To be Published
|
|
6OKE
 
 | |
6ATS
 
 | |
9FMH
 
 | | PsiM N247M in complex with SAH and norbaeocystin | | Descriptor: | CHLORIDE ION, Norbaeocystin, Psilocybin synthase, ... | | Authors: | Hudspeth, J, Rupp, B, Werten, S. | | Deposit date: | 2024-06-06 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The Second Methylation in Psilocybin Biosynthesis Is Enabled by a Hydrogen Bonding Network Extending into the Secondary Sphere Surrounding the Methyltransferase Active Site.
Chembiochem, 25, 2024
|
|
9FMI
 
 | | PsiM N247A in complex with SAH and norbaeocystin | | Descriptor: | CHLORIDE ION, Norbaeocystin, Psilocybin synthase, ... | | Authors: | Hudspeth, J, Rupp, B, Werten, S. | | Deposit date: | 2024-06-06 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The Second Methylation in Psilocybin Biosynthesis Is Enabled by a Hydrogen Bonding Network Extending into the Secondary Sphere Surrounding the Methyltransferase Active Site.
Chembiochem, 25, 2024
|
|
4WKC
 
 | | Crystal structure of Escherichia coli 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TETRAETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
6QB2
 
 | |
5CB8
 
 | |
5CB6
 
 | | Structure of adenosine-5'-phosphosulfate kinase | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Herrmann, J, Jez, J.M. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Recapitulating the Structural Evolution of Redox Regulation in Adenosine 5'-Phosphosulfate Kinase from Cyanobacteria to Plants.
J.Biol.Chem., 290, 2015
|
|
