8OKI
 
 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex bound to Spt4/5 | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8IEJ
 
 | | RNF20-RNF40/hRad6A-Ub/nucleosome complex | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Ai, H, Deng, Z, Sun, M, Du, Y, Pan, M, Liu, L. | | Deposit date: | 2023-02-15 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
9DP1
 
 | | APE1 N174A Substrate Complex with Abasic DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA repair nuclease/redox regulator APEX1, ... | | Authors: | DeHart, K.M, Freudenthal, B.D. | | Deposit date: | 2024-09-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | N174 Physically and Chemically Stabilizes the Endonuclease Reaction of APE1
To Be Published
|
|
4C2M
 
 | | Structure of RNA polymerase I at 2.8 A resolution | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Engel, C, Sainsbury, S, Cheung, A.C, Kostrewa, D, Cramer, P. | | Deposit date: | 2013-08-19 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Polymerase I Structure and Transcription Regulation.
Nature, 502, 2013
|
|
8Q63
 
 | | Cryo-EM structure of IC8', a second state of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-08-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8AP1
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with two GTP molecules poised for de novo initiation (IC2) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATT
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 4-mer RNA, pppGpGpUpA (IC4) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATW
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 6-mer RNA, pppGpGpApApApU (IC6) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
6GYK
 
 | | Structure of a yeast closed complex (core CC1) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Dienemann, C, Schwalb, B, Schilbach, S, Cramer, P. | | Deposit date: | 2018-06-30 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Promoter Distortion and Opening in the RNA Polymerase II Cleft.
Mol. Cell, 73, 2019
|
|
3I4N
 
 | | 8-oxoguanine containing RNA polymerase II elongation complex E | | Descriptor: | DNA (5'-D(*AP*G*CP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP*(8OG)P*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Damsma, G.E, Cramer, P. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Molecular basis of transcriptional mutagenesis at 8-oxoguanine
J.Biol.Chem., 284, 2009
|
|
3I4M
 
 | | 8-oxoguanine containing RNA polymerase II elongation complex D | | Descriptor: | DNA (5'-D(*AP*G*CP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP*(8OG)P*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Damsma, G.E, Cramer, P. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular basis of transcriptional mutagenesis at 8-oxoguanine
J.Biol.Chem., 284, 2009
|
|
6RI7
 
 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RIN
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
3WOD
 
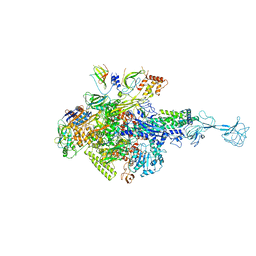 | | RNA polymerase-gp39 complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | Deposit date: | 2013-12-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
8ATV
 
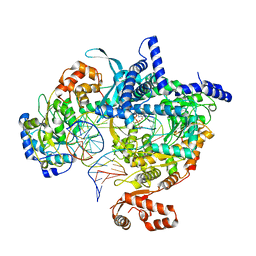 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 5-mer RNA, pppGpGpApApA (IC5) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
5W1T
 
 | |
6YXU
 
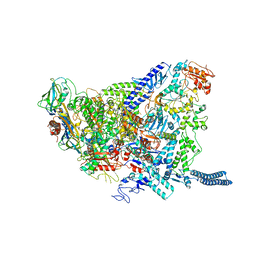 | | Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State I, primary channel engaged | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Koval, T, Krasny, L, Dohnalek, J. | | Deposit date: | 2020-05-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mycobacterial HelD is a nucleic acids-clearing factor for RNA polymerase.
Nat Commun, 11, 2020
|
|
7WBV
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome | | Descriptor: | DNA (159-MER), DNA (198-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBW
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
6YYS
 
 | | Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State II, primary channel engaged and active site interfering | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Koval, T, Krasny, L, Dohnalek, J. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mycobacterial HelD is a nucleic acids-clearing factor for RNA polymerase.
Nat Commun, 11, 2020
|
|
6D6R
 
 | |
6D6Q
 
 | |
1DNT
 
 | | RNA/DNA DODECAMER R(GC)D(GTATACGC) WITH MAGNESIUM BINDING SITES | | Descriptor: | DNA/RNA (5'-R(*GP*CP)-D(*GP*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-16 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
4KMU
 
 | |
6VUG
 
 | |
