3LUC
 
 | |
3LX4
 
 | | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(deltaEFG) | | Descriptor: | ACETATE ION, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Mulder, D.W, Boyd, E.S, Sarma, R, Lange, R.K, Endrizzi, J.A, Broderick, J.B, Peters, J.W. | | Deposit date: | 2010-02-24 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(DeltaEFG).
Nature, 465, 2010
|
|
1R2T
 
 | |
2LKY
 
 | |
1R2B
 
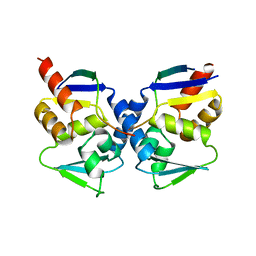 | | Crystal structure of the BCL6 BTB domain complexed with a SMRT co-repressor peptide | | Descriptor: | B-cell lymphoma 6 protein, Nuclear receptor co-repressor 2 | | Authors: | Ahmad, K.F, Melnick, A, Lax, S.A, Bouchard, D, Liu, J, Kiang, C.L, Mayer, S, Licht, J.D, Prive, G.G. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain.
Mol.Cell, 12, 2003
|
|
1R2R
 
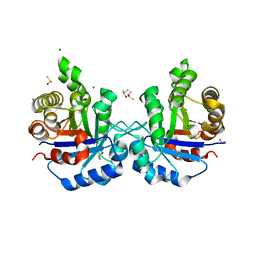 | | CRYSTAL STRUCTURE OF RABBIT MUSCLE TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Aparicio, R, Ferreira, S.T, Polikarpov, I. | | Deposit date: | 2003-09-29 | | Release date: | 2003-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Closed conformation of the active site loop of rabbit muscle triosephosphate isomerase in the absence of substrate: evidence of conformational heterogeneity.
J.Mol.Biol., 334, 2003
|
|
4BF1
 
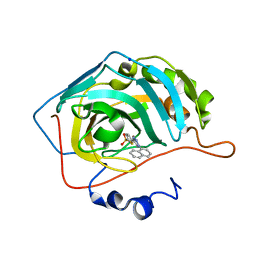 | | Three dimensional structure of human carbonic anhydrase II in complex with 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide | | Descriptor: | 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide, CARBONIC ANHYDRASE 2, SODIUM ION, ... | | Authors: | Tars, K, Leitans, J, Zalubovskis, R. | | Deposit date: | 2013-03-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 5-Substituted-(1,2,3-Triazol-4-Yl)Thiophene-2-Sulfonamides Strongly Inhibit Human Carbonic Anhydrases I, II, Ix and Xii: Solution and X-Ray Crystallographic Studies.
Bioorg.Med.Chem., 21, 2013
|
|
1RI0
 
 | |
3MVD
 
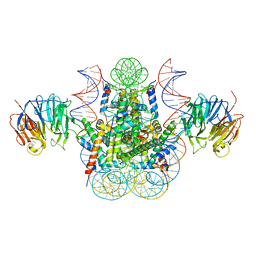 | | Crystal structure of the chromatin factor RCC1 in complex with the nucleosome core particle | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Makde, R.D, England, J.R, Yennawar, H.P, Tan, S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of RCC1 chromatin factor bound to the nucleosome core particle.
Nature, 467, 2010
|
|
1RK8
 
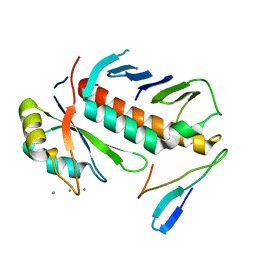 | | Structure of the cytosolic protein PYM bound to the Mago-Y14 core of the exon junction complex | | Descriptor: | CALCIUM ION, CG8781-PA protein, Mago nashi protein, ... | | Authors: | Bono, F, Ebert, J, Guettler, T, Izaurralde, E, Conti, E. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the interaction of PYM with
the Mago-Y14 core of the exon junction complex
EMBO Rep., 5, 2004
|
|
1RFK
 
 | |
1R89
 
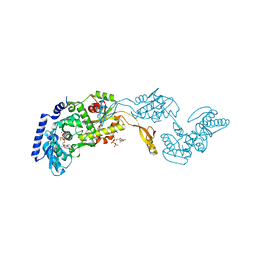 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide Complexes | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
1R4N
 
 | | APPBP1-UBA3-NEDD8, an E1-ubiquitin-like protein complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ubiquitin-like protein NEDD8, ZINC ION, ... | | Authors: | Walden, H, Podgorski, M.S, Holton, J.M, Schulman, B.A. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.
Mol.Cell, 12, 2003
|
|
4BF6
 
 | | Three dimensional structure of human carbonic anhydrase II in complex with 5-(1-(3-Cyanophenyl)-1H-1,2,3-triazol-4-yl)thiophene-2- sulfonamide | | Descriptor: | 5-[1-(3-cyanophenyl)-1,2,3-triazol-4-yl]thiophene-2-sulfonamide, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Tars, K, Leitans, J, Zalubovskis, R. | | Deposit date: | 2013-03-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 5-Substituted-(1,2,3-Triazol-4-Yl)Thiophene-2-Sulfonamides Strongly Inhibit Human Carbonic Anhydrases I, II, Ix and Xii: Solution and X-Ray Crystallographic Studies.
Bioorg.Med.Chem., 21, 2013
|
|
1RAP
 
 | |
3MG9
 
 | | Teg 12 Binary Structure Complexed with the Teicoplanin Aglycone | | Descriptor: | FORMIC ACID, GLYCEROL, TEG12, ... | | Authors: | Bick, M.J, Banik, J.J, Darst, S.A, Brady, S.F. | | Deposit date: | 2010-04-05 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of the Glycopeptide Sulfotransferase Teg12 in a Complex with the Teicoplanin Aglycone.
Biochemistry, 49, 2010
|
|
1R6E
 
 | |
1R8C
 
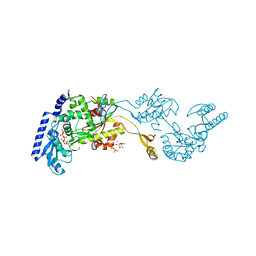 | | Crystal Structures of an Archaeal Class I CCA-Adding Enzyme and Its Nucleotide | | Descriptor: | MANGANESE (II) ION, SODIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Xiong, Y, Li, F, Wang, J, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes
Mol.Cell, 12, 2003
|
|
4BA7
 
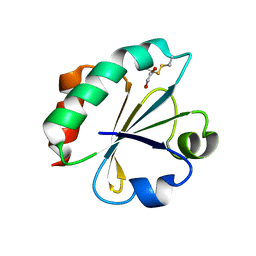 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Bacteria Common Ancestor (LBCA) from the Precambrian Period | | Descriptor: | 1,2-ETHANEDIOL, LBCA THIOREDOXIN | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-09-12 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
1RF6
 
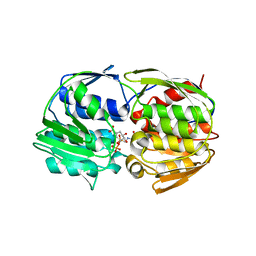 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase in S3P-GLP Bound State | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase, GLYPHOSATE, SHIKIMATE-3-PHOSPHATE | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
6QU1
 
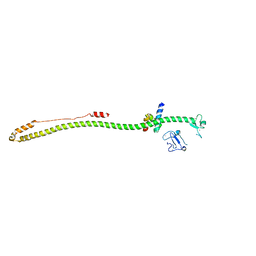 | | Crystal structure of the KAP1 RBCC domain in complex with the SMARCAD1 CUE1 domain at 3.7 angstrom resolution. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1, Transcription intermediary factor 1-beta,Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Newman, J.A, Aitkenhead, H, Gavard, A, Lim, M, Williams, H.L, Svejstrup, J.Q, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A Ubiquitin-Binding Domain that Binds a Structural Fold Distinct from that of Ubiquitin.
Structure, 2019
|
|
1R9Q
 
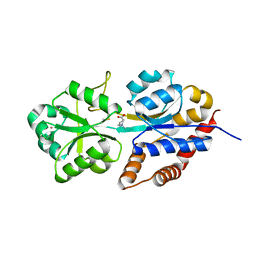 | | structure analysis of ProX in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding periplasmic protein, UNKNOWN ATOM OR ION | | Authors: | Schiefner, A, Breed, J, Bosser, L, Kneip, S, Gade, J, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cation-pi Interactions as Determinants for Binding of the Compatible Solutes Glycine Betaine and Proline Betaine by the Periplasmic Ligand-binding Protein ProX from Escherichia coli
J.BIOL.CHEM., 279, 2004
|
|
3M93
 
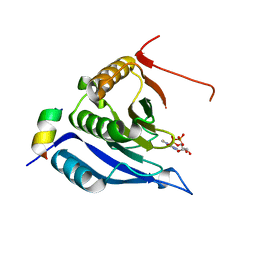 | |
1R4M
 
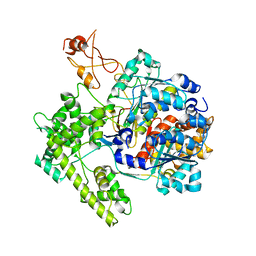 | | APPBP1-UBA3-NEDD8, an E1-ubiquitin-like protein complex | | Descriptor: | Ubiquitin-like protein NEDD8, ZINC ION, amyloid beta precursor protein-binding protein 1, ... | | Authors: | Walden, H, Podgorski, M.S, Holton, J.M, Schulman, B.A. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.
Mol.Cell, 12, 2003
|
|
1RF4
 
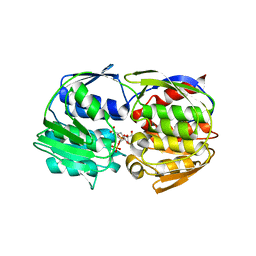 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase, Tetrahedral intermediate Bound State | | Descriptor: | (3R,4S,5R)-5-{[(1R)-1-CARBOXY-2-FLUORO-1-(PHOSPHONOOXY)ETHYL]OXY}-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
