1KK0
 
 | |
1KK1
 
 | | Structure of the large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi-G235D mutant complexed with GDPNP-Mg2+ | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
1KK2
 
 | | Structure of the large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi-G235D mutant complexed with GDP-Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
1KK3
 
 | | Structure of the wild-type large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi complexed with GDP-Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
1KK4
 
 | |
1KK5
 
 | | Crystal Structure of Vat(D) (Form II) | | Descriptor: | STREPTOGRAMIN A ACETYLTRANSFERASE | | Authors: | Sugantino, M, Roderick, S.L. | | Deposit date: | 2001-12-06 | | Release date: | 2002-02-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Vat(D): an acetyltransferase that inactivates streptogramin group A antibiotics.
Biochemistry, 41, 2002
|
|
1KK6
 
 | | Crystal Structure of Vat(D) (Form I) | | Descriptor: | STREPTOGRAMIN A ACETYLTRANSFERASE | | Authors: | Sugantino, M, Roderick, S.L. | | Deposit date: | 2001-12-06 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Vat(D): an acetyltransferase that inactivates streptogramin group A antibiotics.
Biochemistry, 41, 2002
|
|
1KK7
 
 | | SCALLOP MYOSIN IN THE NEAR RIGOR CONFORMATION | | Descriptor: | CALCIUM ION, MAGNESIUM ION, MYOSIN ESSENTIAL LIGHT CHAIN, ... | | Authors: | Himmel, D.M, Gourinath, S, Reshetnikova, L, Shen, Y, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2001-12-06 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KK8
 
 | | SCALLOP MYOSIN (S1-ADP-BeFx) IN THE ACTIN-DETACHED CONFORMATION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Himmel, M, Gourinath, S, Reshetnikova, L, Shen, Y, Szent-Gyorgyi, G, Cohen, C. | | Deposit date: | 2001-12-06 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KK9
 
 | | CRYSTAL STRUCTURE OF E. COLI YCIO | | Descriptor: | SULFATE ION, probable translation factor yciO | | Authors: | Jia, J, Lunin, V.V, Sauve, V, Huang, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-12-06 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the YciO protein from Escherichia coli
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
1KKA
 
 | |
1KKB
 
 | | Complex of Escherichia coli Adenylosuccinate Synthetase with IMP and Hadacidin | | Descriptor: | Adenylosuccinate Synthetase, HADACIDIN, INOSINIC ACID | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-06 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
1KKC
 
 | | Crystal structure of Aspergillus fumigatus MnSOD | | Descriptor: | MANGANESE (II) ION, Manganese Superoxide Dismutase | | Authors: | Fluckiger, S, Mittl, P.R.E, Scapozza, L, Fijten, H, Folkers, G, Grutter, M.G, Blaser, K, Crameri, R. | | Deposit date: | 2001-12-07 | | Release date: | 2001-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparison of the crystal structures of the human manganese superoxide dismutase and the homologous Aspergillus fumigatus allergen at 2-A resolution.
J.Immunol., 168, 2002
|
|
1KKD
 
 | | Solution structure of the calmodulin binding domain (CaMBD) of small conductance Ca2+-activated potassium channels (SK2) | | Descriptor: | Small conductance calcium-activated potassium channel protein 2 | | Authors: | Wissmann, R, Bildl, W, Neumann, H, Rivard, A.F, Kloecker, N, Weitz, D, Schulte, U, Adelman, J.P, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-07 | | Release date: | 2001-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A helical region in the C terminus of small-conductance Ca2+-activated K+ channels controls assembly with apo-calmodulin.
J.Biol.Chem., 277, 2002
|
|
1KKE
 
 | |
1KKF
 
 | | Complex of E. coli Adenylosuccinate Synthetase with IMP, Hadacidin, Pyrophosphate, and Mg | | Descriptor: | Adenylosuccinate Synthetase, DIPHOSPHATE, HADACIDIN, ... | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-07 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
1KKG
 
 | | NMR Structure of Ribosome-Binding Factor A (RbfA) | | Descriptor: | ribosome-binding factor A | | Authors: | Huang, Y.J, Swapna, G.V.T, Rajan, P.K, Ke, H, Xia, B, Shukla, K, Inouye, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-12-07 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ribosome-binding Factor A (RbfA), A Cold-shock
Adaptation Protein from Escherichia coli
J.Mol.Biol., 327, 2003
|
|
1KKH
 
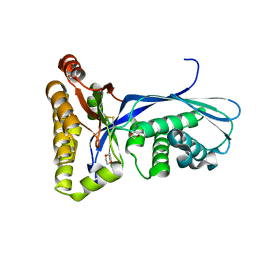 | | Crystal Structure of the Methanococcus jannaschii Mevalonate Kinase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Mevalonate Kinase | | Authors: | Yang, D, Shipman, L.W, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2001-12-08 | | Release date: | 2002-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Methanococcus jannaschii mevalonate kinase, a member of the GHMP kinase superfamily.
J.Biol.Chem., 277, 2002
|
|
1KKJ
 
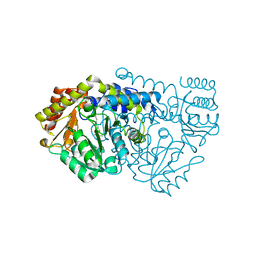 | | Crystal Structure of Serine Hydroxymethyltransferase from B.stearothermophilus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine Hydroxymethyltransferase | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-09 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
1KKK
 
 | |
1KKL
 
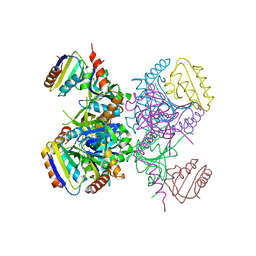 | | L.casei HprK/P in complex with B.subtilis HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHOCARRIER PROTEIN HPR | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KKM
 
 | | L.casei HprK/P in complex with B.subtilis P-Ser-HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHATE ION, ... | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KKO
 
 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE | | Descriptor: | 3-METHYLASPARTATE AMMONIA-LYASE, SULFATE ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, Y, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
1KKP
 
 | | Crystal Structure of Serine Hydroxymethyltransferase complexed with Serine | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, Serine Hydroxymethyltransferase | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
1KKQ
 
 | | Crystal structure of the human PPAR-alpha ligand-binding domain in complex with an antagonist GW6471 and a SMRT corepressor motif | | Descriptor: | N-((2S)-2-({(1Z)-1-METHYL-3-OXO-3-[4-(TRIFLUOROMETHYL) PHENYL]PROP-1-ENYL}AMINO)-3-{4-[2-(5-METHYL-2-PHENYL-1,3-OXAZOL-4-YL)ETHOXY]PHENYL}PROPYL)PROPANAMIDE, NUCLEAR RECEPTOR CO-REPRESSOR 2, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Authors: | Xu, H.E, Stanley, T.B, Montana, V.G, Lambert, M.H, Shearer, B.G, Cobb, J.E, McKee, D.D, Galardi, C.M, Nolte, R.T, Parks, D.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha.
Nature, 415, 2002
|
|
