6X99
 
 | |
6PHK
 
 | |
6PHL
 
 | |
8XIX
 
 | | KeDt3e within Co | | Descriptor: | D-tagatose 3-epimerase | | Authors: | Guo, D. | | Deposit date: | 2023-12-20 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Production of D-psicose
To Be Published
|
|
8XJ1
 
 | | Structure of Apo-KeDt3e | | Descriptor: | D-tagatose 3-epimerase | | Authors: | Guo, D.Y. | | Deposit date: | 2023-12-20 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Production of D-psicose
To Be Published
|
|
7YB3
 
 | | SufS with D-cysteine for 1 min | | Descriptor: | Cysteine desulfurase SufS, D-CYSTEINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing thiazolidine ring formation in the reaction of D-cysteine and pyridoxal-5'-phosphate within L-cysteine desulfurase SufS.
Biochem.Biophys.Res.Commun., 754, 2025
|
|
7X7W
 
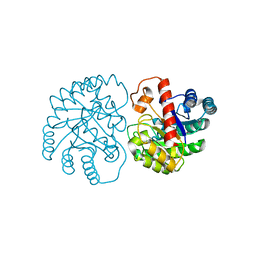 | | The X-ray Crystallographic Structure of D-Psicose 3-epimerase from Clostridia bacterium | | Descriptor: | D-PSICOSE 3-EPIMERASE, MANGANESE (II) ION | | Authors: | Li, Z.F, Ban, X.F, Xie, X.F, Tian, Y.X, Li, C.M, Gu, Z.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of a novel homodimeric D-allulose 3-epimerase from a Clostridia bacterium
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7MSE
 
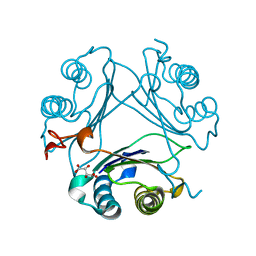 | | High-resolution crystal structure of hMIF2 with tartrate at the active site | | Descriptor: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
6D4L
 
 | |
4WKM
 
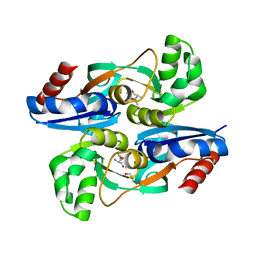 | | AmpR effector binding domain from Citrobacter freundii bound to UDP-MurNAc-pentapeptide | | Descriptor: | ALA-FGA-API-DAL-DAL, GLYCEROL, LysR family transcriptional regulator, ... | | Authors: | Vadlamani, G, Reeve, T.M, Mark, B.L. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The beta-Lactamase Gene Regulator AmpR Is a Tetramer That Recognizes and Binds the d-Ala-d-Ala Motif of Its Repressor UDP-N-acetylmuramic Acid (MurNAc)-pentapeptide.
J.Biol.Chem., 290, 2015
|
|
6D54
 
 | |
6U1C
 
 | |
7C8Q
 
 | | Blasnase-T13A with D-asn | | Descriptor: | Asparaginase, D-ASPARAGINE, FORMIC ACID, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
6N29
 
 | | Crystal structure of monomeric von Willebrand Factor D`D3 assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dong, X, Arndt, J.W, Springer, T.A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The von Willebrand factor D'D3 assembly and structural principles for factor VIII binding and concatemer biogenesis.
Blood, 133, 2019
|
|
3EOV
 
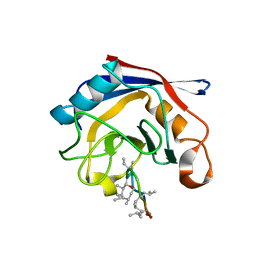 | | Crystal structure of cyclophilin from Leishmania donovani ligated with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Venugopal, V, Dasgupta, D, Datta, A.K, Banerjee, R. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Cyclophilin from Leishmania Donovani Bound to Cyclosporin at 2.6 A Resolution: Correlation between Structure and Thermodynamic Data.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3GPD
 
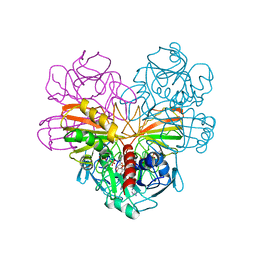 | |
4M6G
 
 | | Structure of the Mycobacterium tuberculosis peptidoglycan amidase Rv3717 in complex with L-Alanine-iso-D-Glutamine reaction product | | Descriptor: | ALANINE, D-alpha-glutamine, Peptidoglycan Amidase Rv3717, ... | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4QE5
 
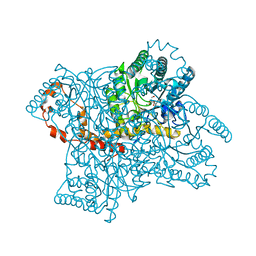 | |
4QE4
 
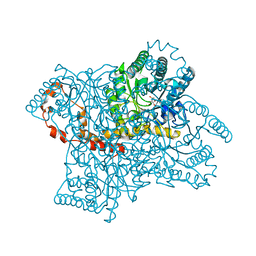 | |
4QEH
 
 | |
2B4K
 
 | | Acetobacter turbidans alpha-amino acid ester hydrolase complexed with phenylglycine | | Descriptor: | Alpha-amino acid ester hydrolase, D-PHENYLGLYCINE, GLYCEROL | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Williams, C, Wybenga, G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-09-26 | | Release date: | 2005-12-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Acetobacter turbidans alpha-amino acid ester hydrolase: how a single mutation improves an antibiotic-producing enzyme.
J.Biol.Chem., 281, 2006
|
|
5ZMY
 
 | | Crystal structure of a cis-epoxysuccinate hydrolase producing D(-)-tartaric acids | | Descriptor: | Cis-epoxysuccinate hydrolase, D(-)-TARTARIC ACID, ZINC ION | | Authors: | Dong, S, Liu, X, Wang, X, Feng, Y. | | Deposit date: | 2018-04-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure d(-)-tartaric acid.
Chem. Commun. (Camb.), 54, 2018
|
|
4QDP
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Cd2+ ions and cyclic beta-L-arabinose | | Descriptor: | CADMIUM ION, Xylose isomerase, beta-L-arabinopyranose | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QE1
 
 | |
4QEE
 
 | |
