4MO3
 
 | |
4KI5
 
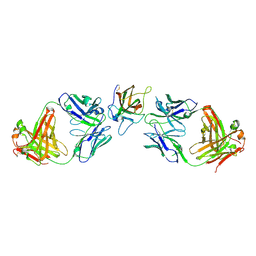 | | Cystal structure of human factor VIII C2 domain in a ternary complex with murine inhbitory antibodies 3E6 and G99 | | Descriptor: | Coagulation factor VIII, MURINE MONOCLONAL 3E6 FAB HEAVY CHAIN, MURINE MONOCLONAL 3E6 FAB LIGHT CHAIN, ... | | Authors: | Walter, J.D, Meeks, S.L, Healey, J.F, Lollar, P, Spiegel, P.C. | | Deposit date: | 2013-05-01 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure of the factor VIII C2 domain in a ternary complex with 2 inhibitor antibodies reveals classical and nonclassical epitopes.
Blood, 122, 2013
|
|
4OUE
 
 | |
4OZO
 
 | |
4LPL
 
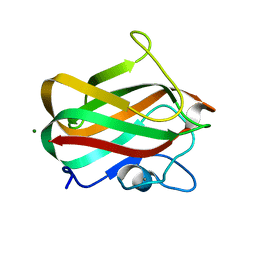 | | Structure of CBM32-1 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein, ... | | Authors: | Grondin, J.M, Duan, D, Heather, F.S, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-16 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
4LQR
 
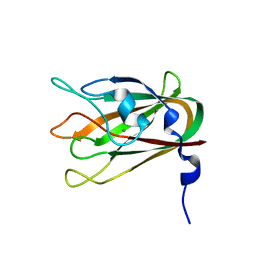 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein | | Authors: | Grondin, J.M, Furness, H.S, Duan, D, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
4LKS
 
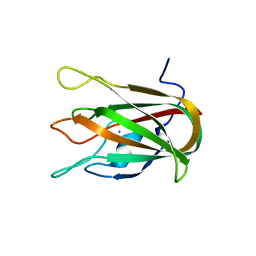 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein, ... | | Authors: | Grondin, J.M, Duan, D, Kirlin, A.C, Furness, H.S, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-08 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
4PT6
 
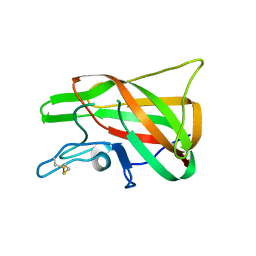 | | The discobody: an engineered discoidin domain from factor VIII that binds v 3 integrin with antibody-like affinities | | Descriptor: | Coagulation factor VIII | | Authors: | Kym, G, Shi, K, Kamajaya, A, Hamdouche, S, Kostecki, J.S, Wannier, T, Li, B, Nikolovski, P, Mayo, S.L. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The discobody: an engineered discoidin domain from factor VIII that binds v 3 integrin with antibody-like affinities
To be Published
|
|
4QDQ
 
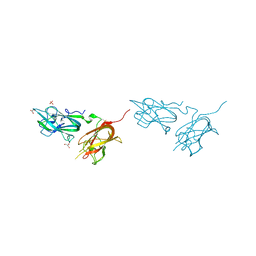 | | Physical basis for Nrp2 ligand binding | | Descriptor: | GLYCEROL, Neuropilin-2, SULFATE ION | | Authors: | Parker, M.W, Vander Kooi, C.W. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for VEGF-C Binding to Neuropilin-2 and Sequestration by a Soluble Splice Form.
Structure, 23, 2015
|
|
4QDR
 
 | |
4QDS
 
 | | Physical basis for Nrp2 ligand binding | | Descriptor: | ACETATE ION, GLYCEROL, Neuropilin-2 | | Authors: | Parker, M.W, Vander Kooi, C.W. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for VEGF-C Binding to Neuropilin-2 and Sequestration by a Soluble Splice Form.
Structure, 23, 2015
|
|
4P5Y
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, Glycosyl hydrolase, ... | | Authors: | Grondin, J.M, Allingham, J.S, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-03-04 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
4RN5
 
 | | B1 domain of human Neuropilin-1 with acetate ion in a ligand-binding site | | Descriptor: | ACETATE ION, GLYCEROL, Neuropilin-1, ... | | Authors: | Allerston, C.K, Yelland, T.S, Jarvis, A, Jenkins, K, Winfield, N, Cheng, L, Jia, H, Zachary, I, Selwood, D.L, Djordjevic, S. | | Deposit date: | 2014-10-23 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Conserved water molecules in a ligand-binding site of neuropilin-1
To be Published
|
|
4TXW
 
 | | Crystal structure of CBM32-4 from the Clostridium perfringens NagH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Hyaluronoglucosaminidase | | Authors: | Grondin, J.M, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-07-07 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution Structure and Dynamics of Full-length GH84A, a multimodular B-N-acetylglucosaminidase from Clostridium perfringens
To Be Published
|
|
4XZU
 
 | |
2RV9
 
 | |
2RVA
 
 | |
4ZZ8
 
 | | X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Glucanase/chitosanase, ... | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZZ5
 
 | | X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZXE
 
 | | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/Chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4ZY9
 
 | | X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | Glucanase/chitosanase | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-21 | | Release date: | 2016-04-13 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
3JD6
 
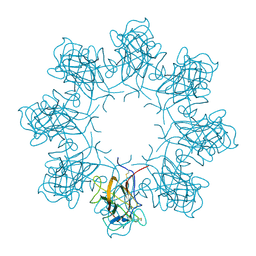 | | Double octamer structure of retinoschisin, a cell-cell adhesion protein of the retina | | Descriptor: | Retinoschisin | | Authors: | Tolun, G, Vijayasarathy, C, Huang, R, Zeng, Y, Li, Y, Steven, A.C, Sieving, P.A, Heymann, J.B. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-11 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Paired octamer rings of retinoschisin suggest a junctional model for cell-cell adhesion in the retina.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5C7G
 
 | |
5DN2
 
 | | Human NRP2 b1 domain in complex with the peptide corresponding to the C-terminus of VEGF-A | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Neuropilin-2, ... | | Authors: | Tsai, Y.C.I, Frankel, P, Fotinou, C, Rana, R, Zachary, I, Djordjevic, S. | | Deposit date: | 2015-09-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of neuropilin-2 reveal a zinc ion binding site remote from the vascular endothelial growth factor binding pocket.
Febs J., 283, 2016
|
|
5K9H
 
 | | Crystal structure of a glycoside hydrolase 29 family member from an unknown rumen bacterium | | Descriptor: | 0940_GH29, GLYCEROL, SODIUM ION, ... | | Authors: | Summers, E.L, Arcus, V.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The structure of a glycoside hydrolase 29 family member from a rumen bacterium reveals unique, dual carbohydrate-binding domains.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
