3DER
 
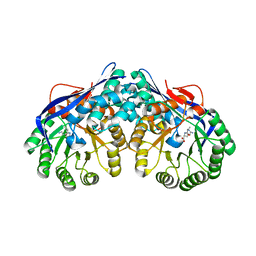 | | Crystal structure of dipeptide epimerase from Thermotoga maritima complexed with L-Ala-L-Lys dipeptide | | Descriptor: | ALANINE, LYSINE, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening.
Structure, 16, 2008
|
|
7S5O
 
 | |
4PG7
 
 | |
4PH1
 
 | |
4PHI
 
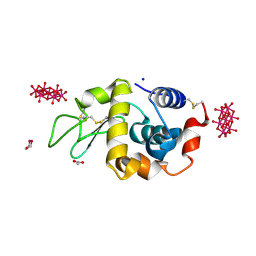 | | Crystal structure of HEWL with hexatungstotellurate(VI) | | Descriptor: | 6-tungstotellurate(VI), ACETATE ION, GLYCEROL, ... | | Authors: | Bijelic, A, Molitor, C, Mauracher, S.G, Al-Oweini, R, Kortz, U, Rompel, A. | | Deposit date: | 2014-05-06 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Hen Egg-White Lysozyme Crystallisation: Protein Stacking and Structure Stability Enhanced by a Tellurium(VI)-Centred Polyoxotungstate.
Chembiochem, 16, 2015
|
|
8I6U
 
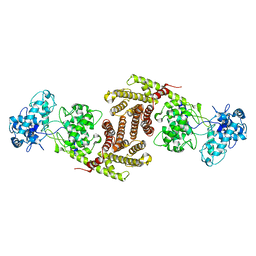 | | The cryo-EM structure of OsCyc1 dimer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8I6T
 
 | | The cryo-EM structure of OsCyc1 hexamer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
4PJM
 
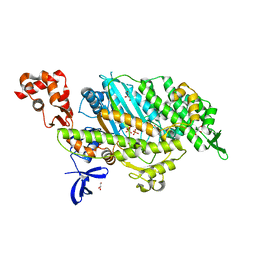 | | Myosin VI motor domain in the Pi release state, space group P212121 - soaked with PO4 - located in the active site | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Isabet, T, Benisty, H, Llinas, P, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | How actin initiates the motor activity of Myosin.
Dev.Cell, 33, 2015
|
|
8I6P
 
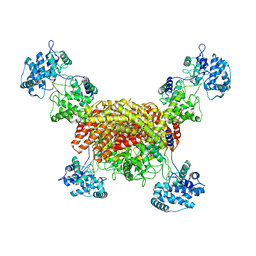 | | The cryo-EM structure of OsCyc1 tetramer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8IH5
 
 | |
3DAE
 
 | |
3DAW
 
 | | Structure of the actin-depolymerizing factor homology domain in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Paavilainen, V.O, Oksanen, E, Goldman, A, Lappalainen, P. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the actin-depolymerizing factor homology domain in complex with actin
J.Cell Biol., 182, 2008
|
|
4PLP
 
 | |
5B6G
 
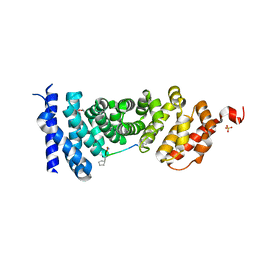 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
4PM9
 
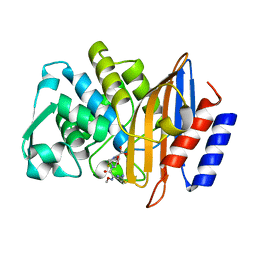 | | Crystal structure of CTX-M-14 S70G:S237A:R276A beta-lactamase in complex with cefotaxime at 1.45 Angstroms resolution | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase CTX-M-14 | | Authors: | Cardenas, A.M, Adamski, C.J, Sankaran, B, Brown, N.G, Horton, L.B, Palzkill, T. | | Deposit date: | 2014-05-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Molecular Basis for the Catalytic Specificity of the CTX-M Extended-Spectrum beta-Lactamases.
Biochemistry, 54, 2015
|
|
1F6R
 
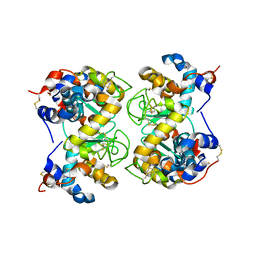 | |
8C89
 
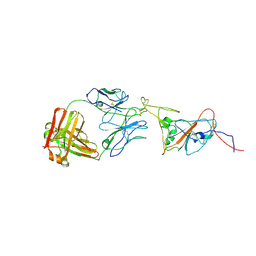 | | SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab) | | Descriptor: | 17T2 Fab heavy chain, 17T2 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Modrego, A, Carlero, D, Bueno-Carrasco, M.T, Santiago, C, Carolis, C, Arranz, R, Blanco, J, Magri, G. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.41 Å) | | Cite: | A monoclonal antibody targeting a large surface of the receptor binding motif shows pan-neutralizing SARS-CoV-2 activity.
Nat Commun, 15, 2024
|
|
7RY1
 
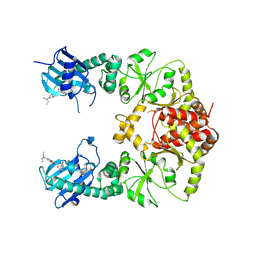 | | human Hsp90_MC domain structure | | Descriptor: | Heat shock protein HSP 90-alpha, N-[2-(1-MALEIMIDYL)ETHYL]-7-DIETHYLAMINOCOUMARIN-3-CARBOXAMIDE | | Authors: | Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-08-24 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.523 Å) | | Cite: | Crystal structure of the middle and C-terminal domains of Hsp90 alpha labeled with a coumarin derivative reveals a potential allosteric binding site as a drug target.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3MLW
 
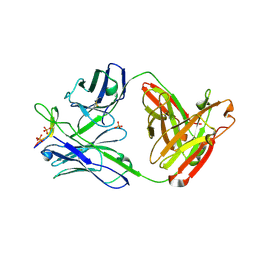 | | Crystal structure of anti-HIV-1 V3 Fab 1006-15D in complex with an MN V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 1006-15D Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 1006-15D Fab light chain, ... | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3DA8
 
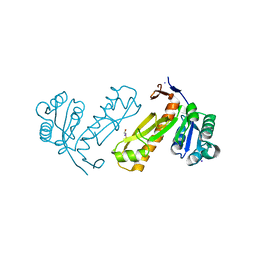 | | Crystal structure of PurN from Mycobacterium tuberculosis | | Descriptor: | BETA-MERCAPTOETHANOL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N. | | Deposit date: | 2008-05-28 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosis reveal a novel dimer with relevance to drug discovery.
J.Mol.Biol., 389, 2009
|
|
2JA4
 
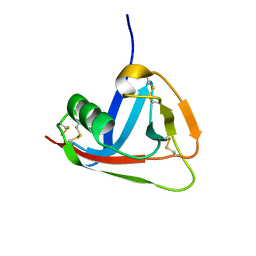 | | Crystal structure of CD5 domain III reveals the fold of a group B scavenger cysteine-rich receptor | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD5 | | Authors: | Rodamilans, B, Munoz, I.G, Sarrias, M.R, Lozano, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2006-11-21 | | Release date: | 2007-03-06 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of the Third Extracellular Domain of Cd5 Reveals the Fold of a Group B Scavenger Cysteine-Rich Receptor Domain.
J.Biol.Chem., 282, 2007
|
|
1F6S
 
 | | CRYSTAL STRUCTURE OF BOVINE ALPHA-LACTALBUMIN | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Chrysina, E.D, Brew, K, Acharya, K.R. | | Deposit date: | 2000-06-23 | | Release date: | 2000-12-13 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of apo- and holo-bovine alpha-lactalbumin at 2. 2-A resolution reveal an effect of calcium on inter-lobe interactions.
J.Biol.Chem., 275, 2000
|
|
3DCJ
 
 | | Crystal structure of glycinamide formyltransferase (PurN) from Mycobacterium tuberculosis in complex with 5-methyl-5,6,7,8-tetrahydrofolic acid derivative | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, ... | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N. | | Deposit date: | 2008-06-03 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosis reveal a novel dimer with relevance to drug discovery.
J.Mol.Biol., 389, 2009
|
|
3DCV
 
 | | Crystal structure of human Pim1 kinase complexed with 4-(4-hydroxy-3-methyl-phenyl)-6-phenylpyrimidin-2(1H)-one | | Descriptor: | 4-(4-hydroxy-3-methylphenyl)-6-phenylpyrimidin-2(5H)-one, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Bellamacina, C.R, Shafer, C.M, Lindvall, M, Gesner, T.G, Yabannavar, A, Weiping, J, Song, L, Walter, A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 4-(1H-indazol-5-yl)-6-phenylpyrimidin-2(1H)-one analogs as potent CDC7 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3MMZ
 
 | | CRYSTAL STRUCTURE OF putative HAD family hydrolase from Streptomyces avermitilis MA-4680 | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative HAD family hydrolase | | Authors: | Malashkevich, V.N, Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
