7YIY
 
 | | Cryo-EM structure of SPT-ORMDL3 complex | | Descriptor: | N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Nat Commun, 14, 2023
|
|
5A19
 
 | | The structure of MAT2A in complex with PPNP. | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2015-04-28 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystallography Captures Catalytic Steps in Human Methionine Adenosyltransferase Enzymes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3M8R
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in a closed ternary complex with trapped 4'-ethylated dTTP | | Descriptor: | 4'-ethylthymidine 5'-(tetrahydrogen triphosphate), ACETATE ION, DNA (5'-D(*AP*AP*AP*AP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Diederichs, K, Marx, A, Betz, K. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of DNA polymerases caught processing size-augmented nucleotide probes.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6K7M
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E2Pi-PL state) | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
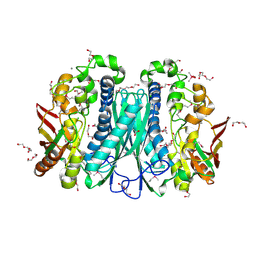
3MDO
 
 | |
7YKA
 
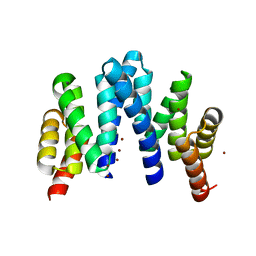 | |
3MMO
 
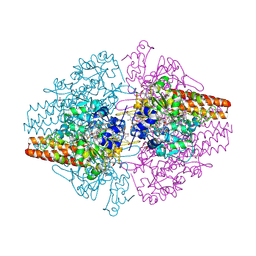 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with cyanide | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-04-20 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of complexes of octahaem cytochrome c nitrite reductase from Thioalkalivibrio nitratireducens with sulfite and cyanide.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6DXY
 
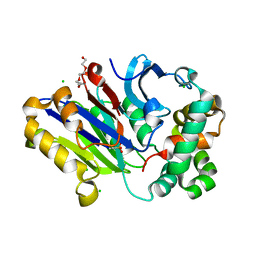 | | Murine N-acylethanolamine-hydrolyzing acid amidase (NAAA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3MSH
 
 | |
5OM3
 
 | | Crystal structure of Alpha1-antichymotrypsin variant DBS-I5: a MMP14-cleavable drug-binding serpin for doxycycline | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, Alpha-1-antichymotrypsin, DI(HYDROXYETHYL)ETHER | | Authors: | Schmidt, K, Muller, Y.A. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of an allosterically modulated doxycycline and doxorubicin drug-binding protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6KM7
 
 | | The structural basis for the internal interaction in RBBP5 | | Descriptor: | HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, Retinoblastoma-binding protein 5, ... | | Authors: | Han, J, Li, T, Chen, Y. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The internal interaction in RBBP5 regulates assembly and activity of MLL1 methyltransferase complex.
Nucleic Acids Res., 47, 2019
|
|
2RBD
 
 | |
5B27
 
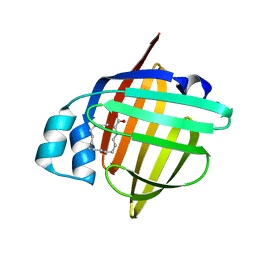 | | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | Authors: | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid.
To Be Published
|
|
5AX0
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.52 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3M8S
 
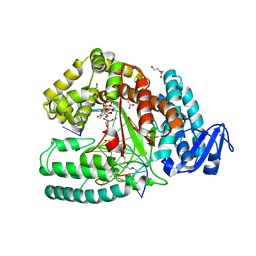 | | Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in a closed ternary complex with trapped 4'-methylated dTTP | | Descriptor: | 4'-methylthymidine 5'-(tetrahydrogen triphosphate), ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Diederichs, K, Marx, A, Betz, K. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of DNA polymerases caught processing size-augmented nucleotide probes.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6DTD
 
 | |
2MCO
 
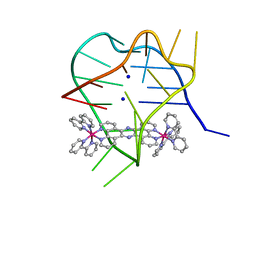 | | Structural studies on dinuclear ruthenium(II) complexes that bind diastereoselectively to an anti-parallel folded human telomere sequence | | Descriptor: | SODIUM ION, human telomere quadruplex, tetrakis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)(mu-tetrapyrido[3,2-a:2',3'-c:3'',2''-h:2''',3'''-j]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~13~,N~14~)diruthenium(4+) L enantiomer | | Authors: | Williamson, M.P, Wilson, T, Thomas, J.A, Felix, V, Costa, P.J. | | Deposit date: | 2013-08-22 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence.
J.Med.Chem., 56, 2013
|
|
3MYG
 
 | | Aurora A Kinase complexed with SCH 1473759 | | Descriptor: | 2-{ethyl[(5-{[6-methyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}isothiazol-3-yl)methyl]amino}-2-methylpropan-1-ol, Serine/threonine-protein kinase 6, TETRAETHYLENE GLYCOL | | Authors: | Hruza, A, Prosis, W, Ramanathan, L. | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Injectable Inhibitor of Aurora Kinases Based on the Imidazo-[1,2-a]-Pyrazine Core.
ACS Med Chem Lett, 1, 2010
|
|
5AOR
 
 | | Structure of MLE RNA ADP AlF4 complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP)-3', ADENOSINE-5'-DIPHOSPHATE, DOSAGE COMPENSATION REGULATOR, ... | | Authors: | Prabu, J.R, Conti, E. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of the RNA Helicase Mle Reveals the Molecular Mechanisms for Uridine Specificity and RNA-ATP Coupling.
Mol.Cell, 60, 2015
|
|
5AOB
 
 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-butyrate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
5DYO
 
 | | Fab43.1 complex with flourescein | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, Fab 43.1 Heavy Chain, Fab 43.1 Light Chain, ... | | Authors: | Longenecker, K.L, Judge, R.A. | | Deposit date: | 2015-09-25 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Three-dimensional structure, binding, and spectroscopic characteristics of the monoclonal antibody 43.1 directed to the carboxyphenyl moiety of fluorescein.
Biopolymers, 105, 2016
|
|
5AWZ
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.57 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5BNM
 
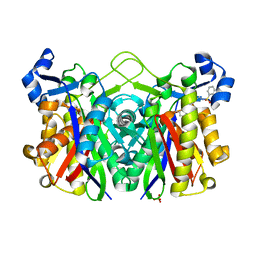 | | E. coli FabH with Small Molecule Inhibitor 1 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, N-{[3'-(hydroxymethyl)biphenyl-4-yl]methyl}benzenesulfonamide, SULFATE ION, ... | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5AMB
 
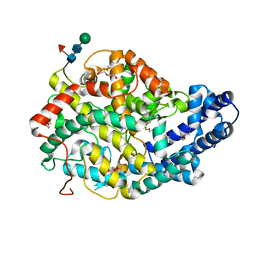 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with amyloid-beta 35-42 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMYLOID BETA A4 PROTEIN, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Masuyer, G, Larmuth, K.M, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Kinetic and Structural Characterisation of Amyloid-Beta Metabolism by Human Angiotensin-1- Converting Enzyme (Ace)
FEBS J., 283, 2016
|
|
5AX1
 
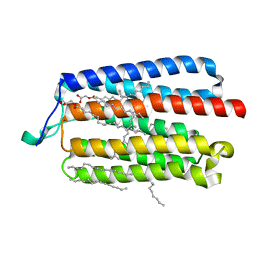 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.80 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
