3TOC
 
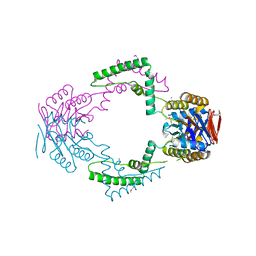 | | Crystal structure of Streptococcus pyogenes Csn2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Putative uncharacterized protein | | Authors: | Bae, E, Jung, D.K, Koo, Y. | | Deposit date: | 2011-09-05 | | Release date: | 2012-05-30 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal Structure of Streptococcus pyogenes Csn2 Reveals Calcium-Dependent Conformational Changes in Its Tertiary and Quaternary Structure
Plos One, 7, 2012
|
|
1YZI
 
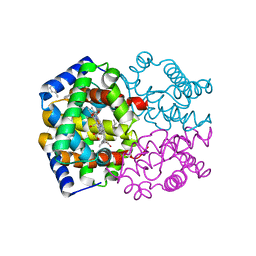 | |
1QV9
 
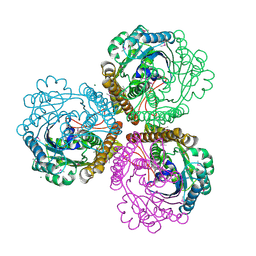 | | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: A methanogenic enzyme with an unusual quarternary structure | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Hagemeier, C.H, Shima, S, Thauer, R.K, Bourenkov, G, Bartunik, H.D, Ermler, U. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: a methanogenic enzyme with an unusual quarternary structure
J.Mol.Biol., 332, 2003
|
|
1QYB
 
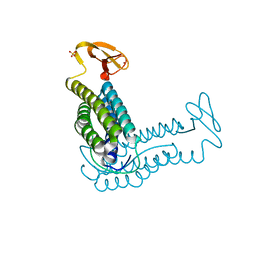 | | X-ray crystal structure of Desulfovibrio vulgaris rubrerythrin with zinc substituted into the [Fe(SCys)4] site and alternative diiron site structures | | Descriptor: | FE (III) ION, Rubrerythrin, SULFATE ION, ... | | Authors: | Jin, S, Kurtz, D.M, Liu, Z.J, Rose, J, Wang, B.C. | | Deposit date: | 2003-09-10 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray Crystal Structure of Desulfovibrio vulgaris Rubrerythrin with Zinc Substituted into the [Fe(SCys)(4)] Site and Alternative Diiron Site Structures.
Biochemistry, 43, 2004
|
|
1NIZ
 
 | | NMR structure of a V3 (MN isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | Exterior membrane glycoprotein(GP120) | | Authors: | Sharon, M, Kessler, N, Levy, R, Zolla-Pazner, S, Gorlach, M, Anglister, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alternative Conformations of HIV-1 V3 Loops Mimic
beta Hairpins in Chemokines, Suggesting a Mechanism
for Coreceptor Selectivity.
Structure, 11, 2003
|
|
6ED2
 
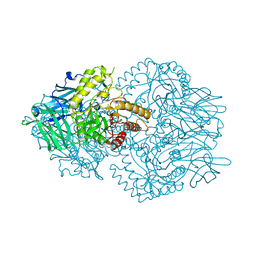 | | Faecalibacterium prausnitzii beta-glucuronidase | | Descriptor: | FORMIC ACID, GLYCEROL, Glycosyl hydrolase family 2, ... | | Authors: | Pellock, S.J, Biernat, K.A, Redinbo, M.R. | | Deposit date: | 2018-08-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
4D9Q
 
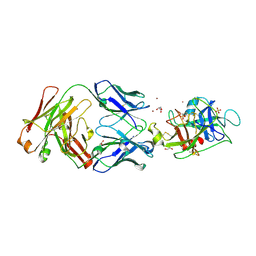 | |
1NJ0
 
 | | NMR structure of a V3 (MN isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | Exterior membrane glycoprotein(GP120) | | Authors: | Sharon, M, Kessler, N, Levy, R, Zolla-Pazner, S, Gorlach, M, Anglister, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alternative Conformations of HIV-1 V3 Loops Mimic
beta Hairpins in Chemokines, Suggesting a Mechanism
for Coreceptor Selectivity.
Structure, 11, 2003
|
|
8PTL
 
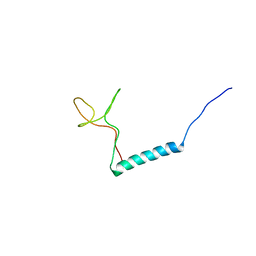 | | human PHOX2B C-terminal domain including the polyA fragment at 278K | | Descriptor: | Paired mesoderm homeobox protein 2B | | Authors: | Anton, R, Trevino, M.A, Pantoja-Uceda, D, Felix, S, Babu, M, Cabrita, E.J, Zweckstetter, M, Tinnefeld, P, Vera, A.M, Oroz, J. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions.
Nat Commun, 15, 2024
|
|
8PUI
 
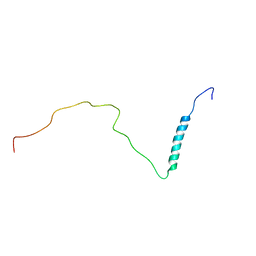 | | human PHOX2B C-terminal domain including the polyA fragment at 298K | | Descriptor: | Paired mesoderm homeobox protein 2B | | Authors: | Anton, R, Trevino, M.A, Pantoja-Uceda, D, Felix, S, Babu, M, Cabrita, E.J, Zweckstetter, M, Tinnefeld, P, Vera, A.M, Oroz, J. | | Deposit date: | 2023-07-17 | | Release date: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions.
Nat Commun, 15, 2024
|
|
4D9R
 
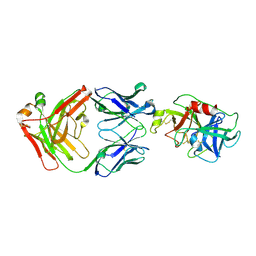 | |
6F38
 
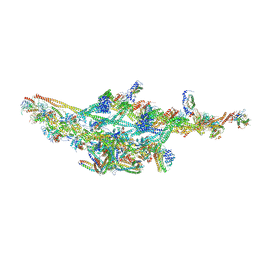 | | Cryo-EM structure of two dynein tail domains bound to dynactin and HOOK3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Urnavicius, L, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
1J9T
 
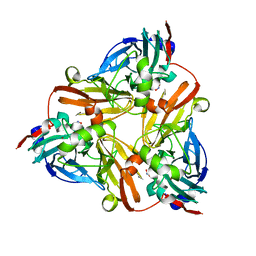 | | Crystal structure of nitrite soaked reduced H255N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
1J9R
 
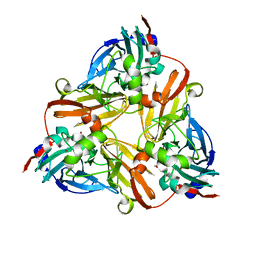 | | Crystal structure of nitrite soaked reduced D98N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
4FF7
 
 | |
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
1J9Q
 
 | | Crystal structure of nitrite soaked oxidized D98N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
1J9S
 
 | | Crystal structure of nitrite soaked oxidized H255N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
4P3X
 
 | | Structure of the Fe4S4 quinolinate synthase NadA from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Cherrier, M.V, Chan, A, Darnault, C, Reichmann, D, Amara, P, Ollagnier de Choudens, S, Fontecilla-Camps, J.C. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of Fe4S4 quinolinate synthase unravels an enzymatic dehydration mechanism that uses tyrosine and a hydrolase-type triad.
J.Am.Chem.Soc., 136, 2014
|
|
4P8I
 
 | | Tgl - a bacterial spore coat transglutaminase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Brito, J.A, Placido, D, Fernandes, C, Lousa, D, Isidro, A, Soares, C.M, Pohl, J, Carrondo, M.A, Henriques, A.O, Archer, M. | | Deposit date: | 2014-03-31 | | Release date: | 2015-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Characterization of an Ancient Bacterial Transglutaminase Sheds Light on the Minimal Requirements for Protein Cross-Linking.
Biochemistry, 54, 2015
|
|
7FJS
 
 | | Crystal structure of T6 Fab bound to theSARS-CoV-2 RBD of B.1.351 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
1NCV
 
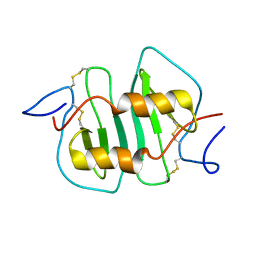 | | DETERMINATION CC-CHEMOKINE MCP-3, NMR, 7 STRUCTURES | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 3 | | Authors: | Meunier, S, Bernassau, J.M, Guillemot, J.C, Ferrara, P, Darbon, H. | | Deposit date: | 1997-02-05 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of CC chemokine monocyte chemoattractant protein 3 by 1H two-dimensional NMR spectroscopy.
Biochemistry, 36, 1997
|
|
2ME3
 
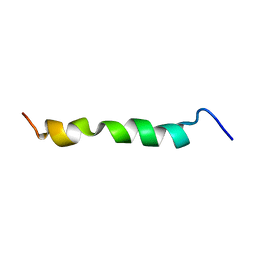 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
4CZP
 
 | |
