2D4A
 
 | | Structure of the malate dehydrogenase from Aeropyrum pernix | | Descriptor: | Malate dehydrogenase | | Authors: | Kawakami, R, Sakuraba, H, Tsuge, H, Ohshima, T. | | Deposit date: | 2005-10-12 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Refolding, characterization and crystal structure of (S)-malate dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
Biochim.Biophys.Acta, 1794, 2009
|
|
7C2V
 
 | | Crystal Structure of IRAK4 kinase in complex with the inhibitor CA-4948 | | Descriptor: | 2-(2-methylpyridin-4-yl)-N-[2-morpholin-4-yl-5-[(3R)-3-oxidanylpyrrolidin-1-yl]-[1,3]oxazolo[4,5-b]pyridin-6-yl]-1,3-oxazole-4-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Krishnamurthy, N.R, Robert, B. | | Deposit date: | 2020-05-09 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of CA-4948, an Orally Bioavailable IRAK4 Inhibitor for Treatment of Hematologic Malignancies.
Acs Med.Chem.Lett., 11, 2020
|
|
7C7Z
 
 | |
7CC4
 
 | |
7CBI
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)pentyl (2~{S},3~{R})-2-azanyl-3-oxidanyl-butanoate, GLYCEROL, ... | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
7CCA
 
 | | Crystal structure of White Spot Syndrome Virus Thymidylate Synthase - ternary complex with Methotrexate and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, METHOTREXATE, Thymidylate Synthase | | Authors: | Panchal, N.V, Kumar, S, Shaikh, N, Vasudevan, D. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure analysis of thymidylate synthase from white spot syndrome virus reveals WSSV-specific structural elements.
Int.J.Biol.Macromol., 167, 2021
|
|
7CFL
 
 | |
7CIC
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 2-azanyl-N-[3-(trifluoromethyloxy)phenyl]ethanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Baker, L.M, Mima, M, Surgenor, A, Robertson, A. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
J.Med.Chem., 63, 2020
|
|
7CIM
 
 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with 3-methlythiopropylamine (geminal diamine form). | | Descriptor: | L-methionine decarboxylase, [6-methyl-4-[(3-methylsulfanylpropylamino)methyl]-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CL5
 
 | | The crystal structure of KanJ in complex with kanamycin B and N-oxalylglycine | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Kanamycin B dioxygenase, N-OXALYLGLYCINE, ... | | Authors: | Kitayama, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2020-07-20 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stepwise Post-glycosylation Modification of Sugar Moieties in Kanamycin Biosynthesis.
Chembiochem, 22, 2021
|
|
2YBK
 
 | | JMJD2A COMPLEXED WITH R-2-HYDROXYGLUTARATE | | Descriptor: | (2R)-2-hydroxypentanedioic acid, CHLORIDE ION, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Chowdhury, R, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-03-08 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep., 12, 2011
|
|
7CU0
 
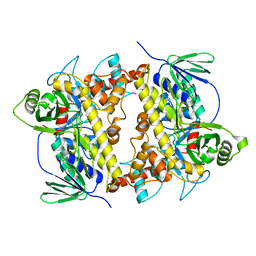 | | Crystal structure of Streptomyces albogriseolus flavin-dependent tryptophan 6-halogenase Thal in complex with tryptophan | | Descriptor: | TRYPTOPHAN, Tryptophan 6-halogenase | | Authors: | Chitnumsub, P, Jaruwat, A, Phintha, A, Chaiyen, P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dissecting the low catalytic capability of flavin-dependent halogenases.
J.Biol.Chem., 296, 2020
|
|
7CW1
 
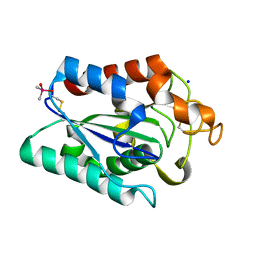 | |
7CWI
 
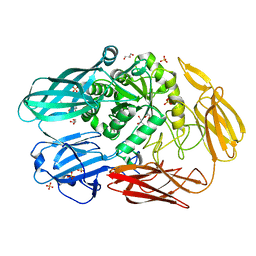 | | Crystal structure of beta-galactosidase II from Bacillus circulans | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Hong, H, Seo, H. | | Deposit date: | 2020-08-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High Galacto-Oligosaccharide Production and a Structural Model for Transgalactosylation of beta-Galactosidase II from Bacillus circulans .
J.Agric.Food Chem., 68, 2020
|
|
7C6A
 
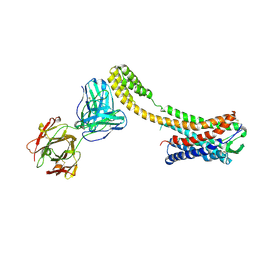 | | Crystal structure of AT2R-BRIL and SRP2070_Fab complex | | Descriptor: | IgG Light Chain, IgG heavy chain, SAR1, ... | | Authors: | Suzuki, M, Miyagi, H, Asada, H, Yasunaga, M, Suno, C, Takahashi, Y, Saito, J, Iwata, S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The discovery of a new antibody for BRIL-fused GPCR structure determination.
Sci Rep, 10, 2020
|
|
7C7Y
 
 | | C-terminal domain of B. cereus TubY | | Descriptor: | Uncharacterized protein | | Authors: | Hayashi, I. | | Deposit date: | 2020-05-27 | | Release date: | 2020-10-28 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The C-terminal region of the plasmid partitioning protein TubY is a tetramer that can bind membranes and DNA.
J.Biol.Chem., 295, 2020
|
|
7CT3
 
 | | Crystal Structure of MglC from Myxococcus xanthus | | Descriptor: | Mutual gliding motility protein C (MglC), SODIUM ION | | Authors: | Thakur, K.G, Kapoor, S, Kodesia, A. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-27 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of Myxococcus xanthus MglC, a component of the polarity control system, and its interactions with its paralog MglB.
J.Biol.Chem., 296, 2021
|
|
7COJ
 
 | | Crystal structure of the b-carbonic anhydrase CafA of the fungal pathogen Aspergillus fumigatus | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Yeon, J, Sung, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of beta-Carbonic Anhydrase CafA from the Fungal Pathogen Aspergillus fumigatus .
Mol.Cells, 43, 2020
|
|
2YKB
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, N-[4-(3H-IMIDAZO[4,5-C]PYRIDIN-2-YL)-9H-FLUOREN-9-YL]-SUCCINAMIDE | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2YKC
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, N-[4-(3H-IMIDAZO[4,5-C]PYRIDIN-2-YL)-9H-FLUOREN-9-YL-ISONICOTINAMIDE | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
7CIG
 
 | | Crystal structure of L-methionine decarboxylase Q64A mutant from Streptomyces sp.590 in complexed with L- methionine methyl ester (geminal diamine form). | | Descriptor: | L-methionine decarboxylase, methyl (2S)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-methylsulfanyl-butanoate | | Authors: | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CP0
 
 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-08-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Piperidine-4-carboxamide as a new scaffold for designing secretory glutaminyl cyclase inhibitors.
Int.J.Biol.Macromol., 170, 2020
|
|
7CQF
 
 | |
7CFO
 
 | | Crystal structure of human RXRalpha ligand binding domain complexed with CBTF-EE. | | Descriptor: | 1-[3-(2-ethoxyethoxy)-5,5,8,8-tetramethyl-6,7-dihydronaphthalen-2-yl]-2-(trifluoromethyl)benzimidazole-5-carboxylic acid, GLYCEROL, Retinoic acid receptor RXR-alpha | | Authors: | Watanabe, M, Fujihara, M, Motoyama, T, Kawasaki, M, Yamada, S, Takamura, Y, Ito, S, Makishima, M, Nakano, S, Kakuta, H. | | Deposit date: | 2020-06-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a "Gatekeeper" Antagonist that Blocks Entry Pathway to Retinoid X Receptors (RXRs) without Allosteric Ligand Inhibition in Permissive RXR Heterodimers.
J.Med.Chem., 64, 2021
|
|
7CUG
 
 | |
