3J4P
 
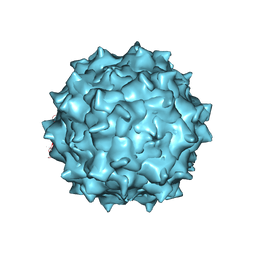 | | Electron Microscopy Analysis of a Disaccharide Analog complex Reveals Receptor Interactions of Adeno-Associated Virus | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Capsid protein VP1, MAGNESIUM ION, ... | | Authors: | Xie, Q, Chapman, M.S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Electron microscopy analysis of a disaccharide analog complex reveals receptor interactions of adeno-associated virus.
J.Struct.Biol., 184, 2013
|
|
4BQC
 
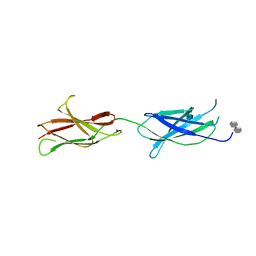 | | Crystal structure of the FN5 and FN6 domains of NEO1 bound to SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN, ... | | Authors: | Bell, C.H, Healey, E, vanErp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
4BOH
 
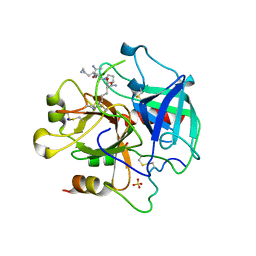 | | Madanins (MEROPS I53) are cleaved by thrombin and factor Xa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, SULFATE ION, ... | | Authors: | Figueiredo, A.C, deSanctis, D, Pereira, P.J.B. | | Deposit date: | 2013-05-20 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | The Tick-Derived Anticoagulant Madanin is Processed by Thrombin and Factor Xa.
Plos One, 8, 2013
|
|
3HYA
 
 | | HYALURONIC ACID, MOLECULAR CONFORMATIONS AND INTERACTIONS IN TWO SODIUM SALTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid, SODIUM ION | | Authors: | Arnott, S. | | Deposit date: | 1977-11-20 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-21 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Hyaluronic acid: molecular conformations and interactions in two sodium salts.
J.Mol.Biol., 95, 1975
|
|
1NT5
 
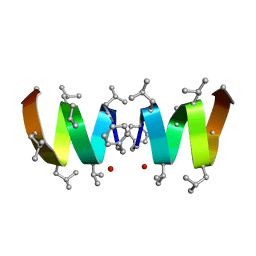 | | F1-Gramicidin A in Sodium Dodecyl Sulfate Micelles (NMR) | | Descriptor: | GRAMICIDIN A | | Authors: | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-11 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
1NNH
 
 | | Hypothetical protein from Pyrococcus furiosus Pfu-1801964 | | Descriptor: | SODIUM ION, asparaginyl-tRNA synthetase-related peptide | | Authors: | Tempel, W, Liu, Z.-J, Schubot, F.D, Shah, A, Arendall III, W.B, Rose, J.P, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-01-13 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hypothetical protein from Pyrococcus furiosus Pfu-1801964
To be published
|
|
1O2G
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-1H-INDOL-2-YL}-1,1'-BIPHENYL-2-OLATE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1O5G
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, CALCIUM ION, Hirudin IIIB', ... | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
3JPY
 
 | | Crystal structure of the zinc-bound amino terminal domain of the NMDA receptor subunit NR2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutamate [NMDA] receptor subunit epsilon-2, ... | | Authors: | Karakas, E, Simorowski, N, Furukawa, H. | | Deposit date: | 2009-09-04 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit.
Embo J., 28, 2009
|
|
3JY9
 
 | | Janus Kinase 2 Inhibitors | | Descriptor: | (3S)-3-(4-hydroxyphenyl)-1,5-dihydro-1,5,12-triazabenzo[4,5]cycloocta[1,2,3-cd]inden-4(3H)-one, SODIUM ION, Tyrosine-protein kinase JAK2 | | Authors: | Zuccola, H.J, Ledeboer, M.W, Pierce, A.C. | | Deposit date: | 2009-09-21 | | Release date: | 2009-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Janus kinase 2 inhibitors. Synthesis and characterization of a novel polycyclic azaindole.
J.Med.Chem., 52, 2009
|
|
4DJ4
 
 | | X-ray structure of mutant N211D of bifunctional nuclease TBN1 from Solanum lycopersicum (Tomato) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Nuclease, ... | | Authors: | Koval, T, Stepankova, A, Lipovova, P, Podzimek, T, Matousek, J, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2012-02-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Plant multifunctional nuclease TBN1 with unexpected phospholipase activity: structural study and reaction-mechanism analysis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1O4Y
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF BETA-AGARASE A FROM ZOBELLIA GALACTANIVORANS | | Descriptor: | CALCIUM ION, SODIUM ION, SULFATE ION, ... | | Authors: | Allouch, J, Jam, M, Helbert, W, Barbeyron, T, Kloareg, B, Henrissat, B, Czjzek, M. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Three-dimensional Structures of Two {beta}-Agarases.
J.Biol.Chem., 278, 2003
|
|
1OB7
 
 | | Cephaibol C | | Descriptor: | CEPHAIBOL C, ETHANOL, SODIUM ION | | Authors: | Bunkoczi, G, Schiell, M, Vertesy, L, Sheldrick, G.M. | | Deposit date: | 2003-01-24 | | Release date: | 2003-12-11 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal Structures of Cephaibols
J.Pept.Sci., 9, 2003
|
|
4DHY
 
 | | Crystal structure of human glucokinase in complex with glucose and activator | | Descriptor: | Glucokinase, N,N-dimethyl-5-({2-methyl-6-[(5-methylpyrazin-2-yl)carbamoyl]-1-benzofuran-4-yl}oxy)pyrimidine-2-carboxamide, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
3HWE
 
 | |
4CNC
 
 | | Crystal structure of human 5T4 (Wnt-activated inhibitory factor 1, Trophoblast glycoprotein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhao, Y, Malinauskas, T, Harlos, K, Jones, E.Y. | | Deposit date: | 2014-01-21 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Insights Into the Inhibition of Wnt Signaling by Cancer Antigen 5T4/Wnt-Activated Inhibitory Factor 1.
Structure, 22, 2014
|
|
4CUO
 
 | | Banyan peroxidase with glycosylation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BANYAN PEROXIDASE, CALCIUM ION, ... | | Authors: | Palm, G.J, Sharma, A, Hinrichs, W. | | Deposit date: | 2014-03-20 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Post-Translational Modification and Extended Glycosylation Pattern of a Plant Latex Peroxidase of Native Source Characterized by X-Ray Crystallography.
FEBS J., 281, 2014
|
|
4F9O
 
 | |
1OXU
 
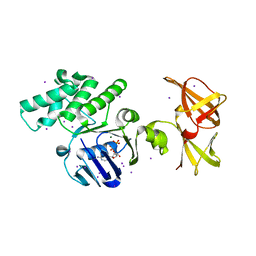 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Verdon, G, Albers, S.V, Dijkstra, B.W, Driessen, A.J, Thunnissen, A.M. | | Deposit date: | 2003-04-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations
J.Mol.Biol., 330, 2003
|
|
4FOI
 
 | |
4FPA
 
 | |
1QSX
 
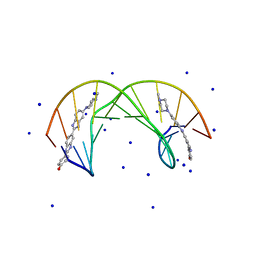 | | SOLUTION NMR STRUCTURE OF THE 2:1 HOECHST 33258-D(CTTTTGCAAAAG)2 COMPLEX | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(CP*TP*TP*TP*TP*GP*CP*AP*AP*AP*AP*G)-3', SODIUM ION | | Authors: | Gavathiotis, E, Sharman, G.J, Searle, M.S. | | Deposit date: | 1999-06-24 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sequence-dependent variation in DNA minor groove width dictates orientational preference of Hoechst 33258 in A-tract recognition: solution NMR structure of the 2:1 complex with d(CTTTTGCAAAAG)(2).
Nucleic Acids Res., 28, 2000
|
|
4DXO
 
 | |
1Q9I
 
 | | The A251C:S430C double mutant of flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HEME C, MALATE LIKE INTERMEDIATE, ... | | Authors: | Rothery, E.L, Mowat, C.G, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing Domain Mobility in a Flavocytochrome
Biochemistry, 42, 2004
|
|
1QTK
 
 | | CRYSTAL STRUCTURE OF HEW LYSOZYME UNDER PRESSURE OF KRYPTON (55 BAR) | | Descriptor: | CHLORIDE ION, KRYPTON, LYSOZYME, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-06-28 | | Release date: | 1999-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
