2VBG
 
 | | The complex structure of the branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis with 2R-1-hydroxyethyl-deazaThDP | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, BRANCHED-CHAIN ALPHA-KETOACID DECARBOXYLASE, MAGNESIUM ION | | Authors: | Berthold, C.L, Gocke, D, Wood, M.D, Leeper, F, Pohl, M, Schneider, G. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Branched-Chain Keto Acid Decarboxylase (Kdca) from Lactococcus Lactis Provides Insights Into the Structural Basis for the Chemo- and Enantioselective Carboligation Reaction
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3ITJ
 
 | | Crystal structure of Saccharomyces cerevisiae thioredoxin reductase 1 (Trr1) | | Descriptor: | CITRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase 1 | | Authors: | Oliveira, M.A, Discola, K.F, Alves, S.V, Medrano, F.J, Guimaraes, B.G, Netto, L.E.S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the specificity of thioredoxin reductase-thioredoxin interactions. A structural and functional investigation of the yeast thioredoxin system.
Biochemistry, 49, 2010
|
|
4KZ5
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 5 (N-{[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}glycine) | | Descriptor: | Beta-lactamase, N-{[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}glycine, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
3IOZ
 
 | |
1SSA
 
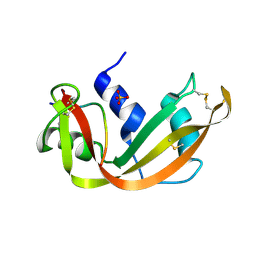 | | A STRUCTURAL INVESTIGATION OF CATALYTICALLY MODIFIED F12OL AND F12OY SEMISYNTHETIC RIBONUCLEASES | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | deMel, V.S.J, Doscher, M.S, Glinn, M.A, Martin, P.D, Ram, M.L, Edwards, B.F.P. | | Deposit date: | 1993-08-03 | | Release date: | 1994-09-30 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of catalytically modified F120L and F120Y semisynthetic ribonucleases.
Protein Sci., 3, 1994
|
|
4FLN
 
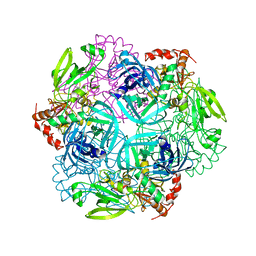 | | Crystal structure of plant protease Deg2 | | Descriptor: | Protease Do-like 2, chloroplastic, Unknown peptide | | Authors: | Gong, W, Liu, L, Sun, R, Gao, F. | | Deposit date: | 2012-06-15 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Arabidopsis deg2 protein reveals an internal PDZ ligand locking the hexameric resting state.
J.Biol.Chem., 287, 2012
|
|
7X2F
 
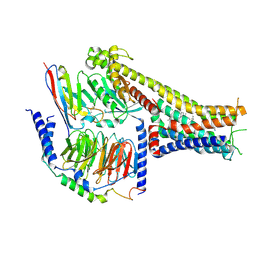 | | Cryo-EM structure of the dopamine and LY3154207-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
1VDY
 
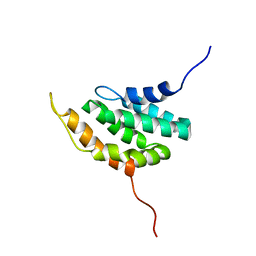 | | NMR Structure of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana | | Descriptor: | hypothetical protein (RAFL09-17-B18) | | Authors: | Lopez-Mendez, B, Pantoja-Uceda, D, Tomizawa, T, Koshiba, S, Kigawa, T, Shirouzu, M, Terada, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical ENTH-VHS domain AT3G16270 from arabidopsis thaliana
To be Published
|
|
5H59
 
 | | Ferredoxin-NADP+ reductase from maize root | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Kurisu, G, Hase, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP(+) oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation
Photosyn. Res., 134, 2017
|
|
7X2C
 
 | | Cryo-EM structure of the fenoldopam-bound D1 dopamine receptor and mini-Gs complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Teng, X, Zheng, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and biased agonism of the D1 dopamine receptor.
Nat Commun, 13, 2022
|
|
1SA0
 
 | | TUBULIN-COLCHICINE: STATHMIN-LIKE DOMAIN COMPLEX | | Descriptor: | 2-MERCAPTO-N-[1,2,3,10-TETRAMETHOXY-9-OXO-5,6,7,9-TETRAHYDRO-BENZO[A]HEPTALEN-7-YL]ACETAMIDE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ravelli, R.B, Gigant, B, Curmi, P.A, Jourdain, I, Lachkar, S, Sobel, A, Knossow, M. | | Deposit date: | 2004-02-06 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain.
Nature, 428, 2004
|
|
6CVH
 
 | | Identification and biological evaluation of thiazole-based inverse agonists of RORgt | | Descriptor: | Nuclear receptor ROR-gamma, trans-3-({4-(cyclohexylmethyl)-5-[3-(1-methylcyclopropyl)-5-{[(2R)-1,1,1-trifluoropropan-2-yl]carbamoyl}phenyl]-1,3-thiazole-2-carbonyl}amino)cyclobutane-1-carboxylic acid | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification and biological evaluation of thiazole-based inverse agonists of ROR gamma t.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
4KZ4
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 60 (2-[(propylsulfonyl)amino]benzoic acid) | | Descriptor: | 2-[(propylsulfonyl)amino]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4LMU
 
 | | Crystal structure of Pim1 in complex with the inhibitor Quercetin (resulting from displacement of SKF86002) | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1VEE
 
 | | NMR structure of the hypothetical rhodanese domain At4g01050 from Arabidopsis thaliana | | Descriptor: | proline-rich protein family | | Authors: | Pantoja-Uceda, D, Lopez-Mendez, B, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Tanaka, A, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the rhodanese homology domain At4g01050(175-295) from Arabidopsis thaliana
Protein Sci., 14, 2005
|
|
3QMM
 
 | |
4JFX
 
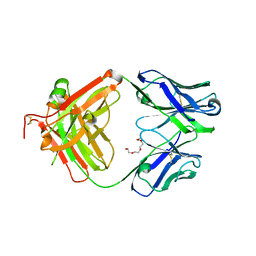 | | Structure of phosphotyrosine (pTyr) scaffold bound to pTyr peptide | | Descriptor: | Fab heavy chain, Fab light chain, Phosphopeptide, ... | | Authors: | Koerber, J.T, Thomsen, N.D, Hannigan, B.T, Degrado, W.F, Wells, J.A. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nature-inspired design of motif-specific antibody scaffolds.
Nat.Biotechnol., 31, 2013
|
|
5OJ9
 
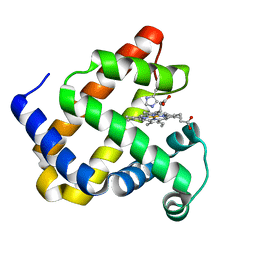 | | Structure of Mb NMH | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
4NR7
 
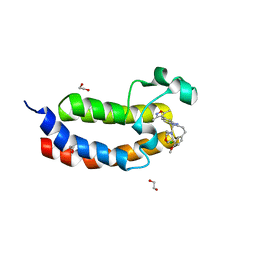 | | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, Krojer, T, Nowak, R, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand
To be Published
|
|
3W51
 
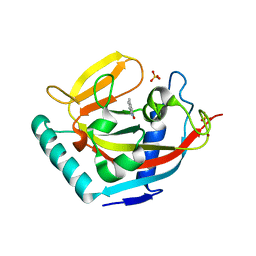 | | Tankyrase in complex with 2-hydroxy-4-methylquinoline | | Descriptor: | 4-methylquinolin-2-ol, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-01-18 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
3WSA
 
 | | The Tuberculosis Drug SQ109 Inhibits Trypanosoma cruzi Cell Proliferation and acts Synergistically with Posaconazole | | Descriptor: | N-[(2Z)-3,7-dimethylocta-2,6-dien-1-yl]-N'-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]ethane-1,2-diamine, Squalene synthase | | Authors: | Shang, N, Li, Q, Huang, C.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | SQ109, a new drug lead for Chagas disease.
Antimicrob.Agents Chemother., 59, 2015
|
|
4LUD
 
 | | Crystal Structure of HCK in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5AHU
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-1326 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, PUTATIVE, MAGNESIUM ION, ... | | Authors: | Yang, G, Oldfield, E, No, J.H. | | Deposit date: | 2015-02-09 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Inhibition of Trypanosoma Brucei Cell Growth by Lipophilic Bisphosphonates: An in Vitro and in Vivo Investigation.
Antimicrob.Agents Chemother., 59, 2015
|
|
6AZJ
 
 | | Crystal Structure of human NAMPT in complex with NVP-LQN520 | | Descriptor: | (1S,2S)-N-{4-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]phenyl}-2-(pyridin-3-yl)cyclopropane-1-carboxamide, Nicotinamide phosphoribosyltransferase | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-09-11 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
6B75
 
 | | Crystal Structure of human NAMPT in complex with NVP-LOQ594 | | Descriptor: | 4-[(piperazin-1-yl)methyl]-N-{[4-({[(pyridin-3-yl)methyl]carbamoyl}amino)phenyl]methyl}benzamide, Nicotinamide phosphoribosyltransferase | | Authors: | Weihofen, W.A, Thigale, S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification and structure based design of cellularly active cyclo-propyl carboxamide Nicotinamide phosphoribosyltransferase (NAMPT) inhibitors
To Be Published
|
|
