6OOB
 
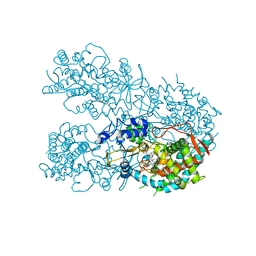 | | Human CYP3A4 bound to a suicide substrate | | Descriptor: | 4-{[(2Z,6S)-6,7-dihydroxy-3,7-dimethyloct-2-en-1-yl]oxy}-7H-furo[3,2-g][1]benzopyran-7-one, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Insights into the Interaction of Cytochrome P450 3A4 with Suicide Substrates: Mibefradil, Azamulin and 6',7'-Dihydroxybergamottin.
Int J Mol Sci, 20, 2019
|
|
5HDR
 
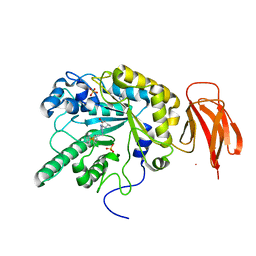 | | Crystal structure of a bacterial fucosidase with iminocyclitol (2S,3S,4R,5S)-3,4-dihydroxy-2-ethynyl-5-methylpyrrolidine | | Descriptor: | Alpha-L-fucosidase, IMIDAZOLE, SULFATE ION, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2016-01-05 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial fucosidase with iminocyclitol (2S,3S,4R,5S)-3,4-dihydroxy-2-ethynyl-5-methylpyrrolidine
To Be Published
|
|
6INH
 
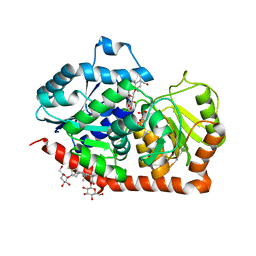 | | A glycosyltransferase with UDP and the substrate | | Descriptor: | 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, UDP-glycosyltransferase 76G1, ... | | Authors: | Zhu, X. | | Deposit date: | 2018-10-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
5HG5
 
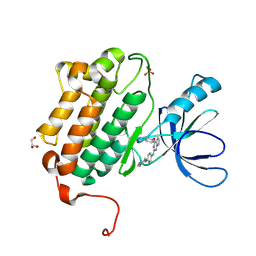 | | EGFR (L858R, T790M, V948R) in complex with N-{3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, GLYCEROL, N-{3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}propanamide, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
7MPH
 
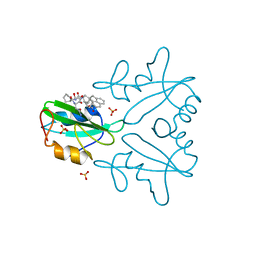 | | GRB2 SH2 Domain with Compound 7 | | Descriptor: | (4-{(10R,11E,14S,18S)-18-(2-amino-2-oxoethyl)-14-[(naphthalen-1-yl)methyl]-8,17,20-trioxo-7,16,19-triazaspiro[5.14]icos-11-en-10-yl}phenyl)acetic acid, 1,2-ETHANEDIOL, Growth factor receptor-bound protein 2, ... | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structural characterization of a monocarboxylic inhibitor for GRB2 SH2 domain.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
6IES
 
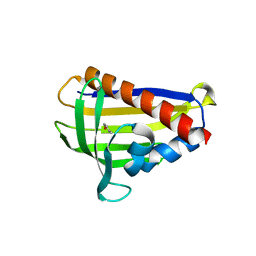 | | Onion lachrymatory factor synthase (LFS) containing (E)-2-propen 1-ol (crotyl alcohol) | | Descriptor: | (2E)-but-2-en-1-ol, Lachrymatory-factor synthase | | Authors: | Sato, Y, Arakawa, T, Takabe, J, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase.
Acs Catalysis, 10, 2020
|
|
6OHT
 
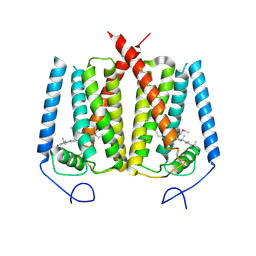 | | Structure of EBP and U18666A | | Descriptor: | 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase, 3beta-(2-Diethylaminoethoxy)androst-5-en-17-one | | Authors: | Long, T, Li, X. | | Deposit date: | 2019-04-06 | | Release date: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for human sterol isomerase in cholesterol biosynthesis and multidrug recognition.
Nat Commun, 10, 2019
|
|
5HWG
 
 | |
3QQK
 
 | | CDK2 in complex with inhibitor L4 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](phenyl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
3QN1
 
 | | Crystal structure of the PYR1 Abscisic Acid receptor in complex with the HAB1 type 2C phosphatase catalytic domain | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Marquez, J.A. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of Abscisic Acid Signaling in Vivo by an Engineered Receptor-Insensitive Protein Phosphatase Type 2C Allele.
Plant Physiol., 156, 2011
|
|
3QLH
 
 | | HIV-1 Reverse Transcriptase in Complex with Manicol at the RNase H Active Site and TMC278 (Rilpivirine) at the NNRTI Binding Pocket | | Descriptor: | (2S)-5,7-dihydroxy-9-methyl-2-(prop-1-en-2-yl)-1,2,3,4-tetrahydro-6H-benzo[7]annulen-6-one, 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Himmel, D.M, Wojtak, K, Bauman, J.D, Arnold, E. | | Deposit date: | 2011-02-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis, activity, and structural analysis of novel alpha-hydroxytropolone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H.
J.Med.Chem., 54, 2011
|
|
6IB0
 
 | | The structure of MKK7 in complex with the covalent 4-amino-pyrazolopyrimidine 3a | | Descriptor: | 1-[(3~{R})-3-(4-azanyl-3-ethynyl-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl]prop-2-en-1-one, Dual specificity mitogen-activated protein kinase kinase 7, TETRAETHYLENE GLYCOL | | Authors: | Wolle, P, Hardick, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting the MKK7-JNK (Mitogen-Activated Protein Kinase Kinase 7-c-Jun N-Terminal Kinase) Pathway with Covalent Inhibitors.
J.Med.Chem., 62, 2019
|
|
3RI5
 
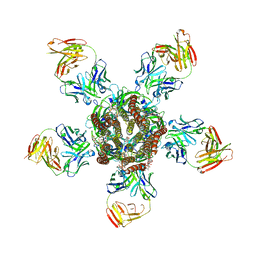 | | C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and picrotoxin | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hibbs, R.E, Gouaux, E. | | Deposit date: | 2011-04-12 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Principles of activation and permeation in an anion-selective Cys-loop receptor.
Nature, 474, 2011
|
|
6OLX
 
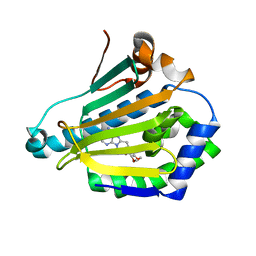 | | Hsp90-alpha S52A bound to PU-11-trans | | Descriptor: | 9-[(2E)-but-2-en-1-yl]-8-[(3,4,5-trimethoxyphenyl)methyl]-9H-purin-6-amine, Heat shock protein HSP 90-alpha | | Authors: | Gewirth, D.T, Huck, J.D. | | Deposit date: | 2019-04-17 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43760586 Å) | | Cite: | Structures of Hsp90 alpha and Hsp90 beta bound to a purine-scaffold inhibitor reveal an exploitable residue for drug selectivity.
Proteins, 87, 2019
|
|
6OMU
 
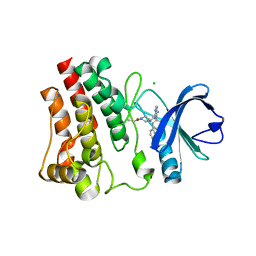 | | Structure of human Bruton's Tyrosine Kinase in complex with Evobrutinib | | Descriptor: | 1-[4-({[6-amino-5-(4-phenoxyphenyl)pyrimidin-4-yl]amino}methyl)piperidin-1-yl]prop-2-en-1-one, CHLORIDE ION, Tyrosine-protein kinase BTK | | Authors: | Mochalkin, I, Gardberg, A.S. | | Deposit date: | 2019-04-19 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Discovery of Evobrutinib: An Oral, Potent, and Highly Selective, Covalent Bruton's Tyrosine Kinase (BTK) Inhibitor for the Treatment of Immunological Diseases.
J.Med.Chem., 62, 2019
|
|
6OAR
 
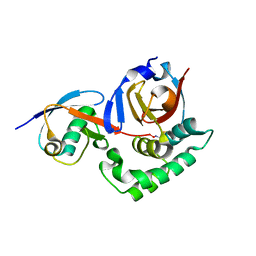 | |
6OAT
 
 | | Structure of the Ganjam virus OTU bound to sheep ISG15 | | Descriptor: | Interferon stimulated gene 17, RNA-dependent RNA polymerase, prop-2-en-1-amine | | Authors: | Dzimianski, J.V, Williams, I.L, Pegan, S.D. | | Deposit date: | 2019-03-18 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Determining the molecular drivers of species-specific interferon-stimulated gene product 15 interactions with nairovirus ovarian tumor domain proteases.
Plos One, 14, 2019
|
|
3REO
 
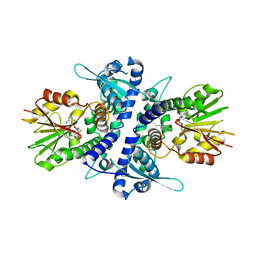 | | Monolignol O-methyltransferase (MOMT) | | Descriptor: | (Iso)eugenol O-methyltransferase, 2-methoxy-4-[(1E)-prop-1-en-1-yl]phenol, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bhuiya, M.W, Liu, C.J. | | Deposit date: | 2011-04-04 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Monolignol O-methyltransferase (MOMT)
To be Published
|
|
5I1Q
 
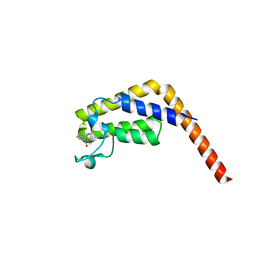 | | Second bromodomain of TAF1 bound to a pyrrolopyridone compound | | Descriptor: | 3-[6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl]-N,N-dimethylbenzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Poy, F, Bellon, S.F. | | Deposit date: | 2016-01-09 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
6OWE
 
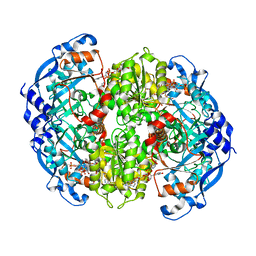 | | Enoyl-CoA carboxylases/reductases in complex with ethylmalonyl CoA | | Descriptor: | 5'-O-[(S)-{[(S)-[(3R)-4-({(1E)-3-[(2-{[(2S)-2-carboxybutanoyl]sulfanyl}ethyl)amino]-3-oxoprop-1-en-1-yl}amino)-3-hydroxy-2,2-dimethyl-4-oxobutoxy](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]adenosine 3'-(dihydrogen phosphate), Crotonyl-CoA carboxylase/reductase, IMIDAZOLE, ... | | Authors: | DeMirci, H. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Four amino acids define the CO2binding pocket of enoyl-CoA carboxylases/reductases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OWC
 
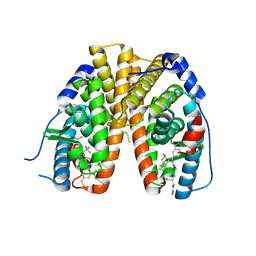 | | Mutant estrogen receptor alpha (ERa) Y537S covalently bound to H3B-6545. | | Descriptor: | (2Z)-N,N-dimethyl-4-{[2-({5-[(1Z)-4,4,4-trifluoro-1-(3-fluoro-2H-indazol-5-yl)-2-phenylbut-1-en-1-yl]pyridin-2-yl}oxy)ethyl]amino}but-2-enamide, 1,2-ETHANEDIOL, Estrogen receptor | | Authors: | Larsen, N.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mutant estrogen receptor alpha (ERa) Y537S covalently bound to H3B-6545.
To Be Published
|
|
5YD6
 
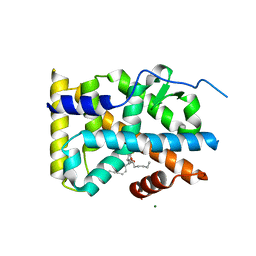 | | Crystal structure of PG-bound Nurr1-LBD | | Descriptor: | (~{Z})-7-[(1~{R},5~{S})-2-oxidanylidene-5-[(~{E},3~{S})-3-oxidanyloct-1-enyl]cyclopent-3-en-1-yl]hept-5-enoic acid, MAGNESIUM ION, Nuclear receptor subfamily 4 group A member 2 | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2017-09-11 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of Nurr1 bound to cyclopentenone prostaglandin A2 and its mechanism of action in ameliorating dopaminergic neurodegeneration in Drosophila
To Be Published
|
|
3QTQ
 
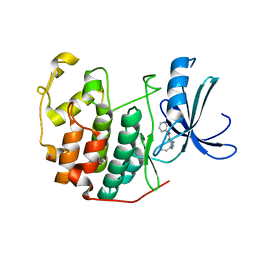 | | CDK2 in complex with inhibitor RC-1-137 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](pyridin-3-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
6JAV
 
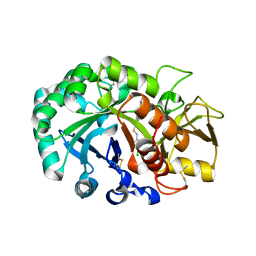 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with a piperidine-thienopyridine derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-chlorophenyl)methyl]sulfanyl}-7-methyl-N-(prop-2-en-1-yl)-7,8-dihydropyrido[4',3':4,5]thieno[2,3-d]pyrimidin-4-amine, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.437 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
