2KAF
 
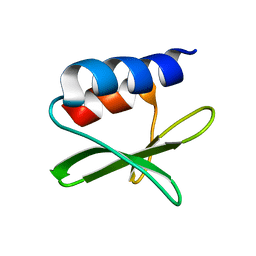 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | Descriptor: | Non-structural protein 3 | | Authors: | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
7R3N
 
 | |
6E76
 
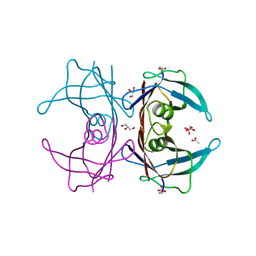 | | Structure of Human Transthyretin Asp38Ala/Thr119Met Mutant | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Saelices, L, Chung, K, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
6E78
 
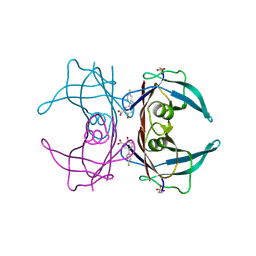 | | Structure of Human Transthyretin Asp38Ala Mutant in Complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, Transthyretin | | Authors: | Chung, K, Saelices, L, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | Descriptor: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
1H4J
 
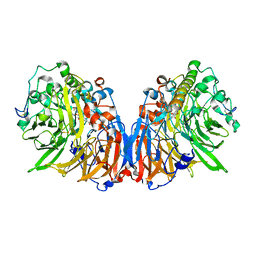 | | Methylobacterium extorquens methanol dehydrogenase D303E mutant | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 1, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Mohammed, F, Gill, R, Thompson, D, Cooper, J.B, Wood, S.P, Afolabi, P.R, Anthony, C. | | Deposit date: | 2001-05-11 | | Release date: | 2001-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Site-Directed Mutagenesis and X-Ray Crystallography of the Pqq-Containing Quinoprotein Methanol Dehydrogenase and its Electron Acceptor, Cytochrome C(L)(,)
Biochemistry, 40, 2001
|
|
3EKE
 
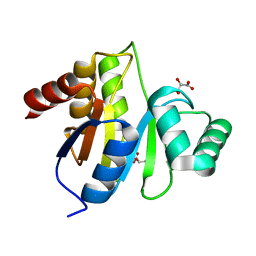 | | Crystal structure of IBV X-domain at pH 5.6 | | Descriptor: | L(+)-TARTARIC ACID, Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
5MQX
 
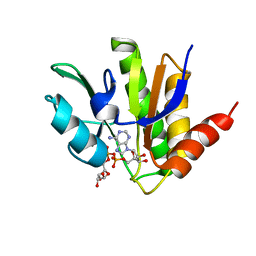 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus(VEEV) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein3 | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Matsoukas, M.T, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
8EVM
 
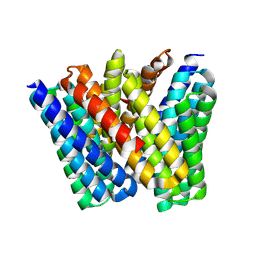 | |
3EJG
 
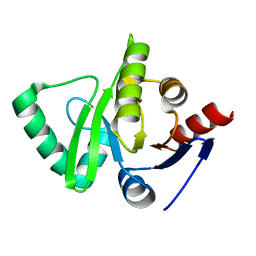 | | Crystal structure of HCoV-229E X-domain | | Descriptor: | Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-18 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
6YXJ
 
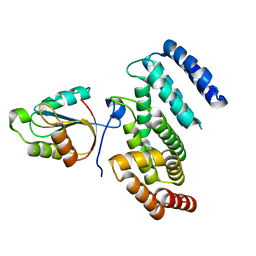 | |
6ZIF
 
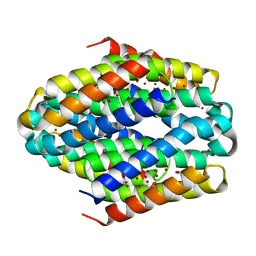 | |
7UV5
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Papain-like protease nsp3, Ubiquitin, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-29 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
1ONS
 
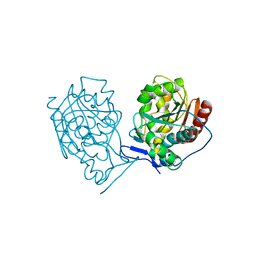 | |
6LNW
 
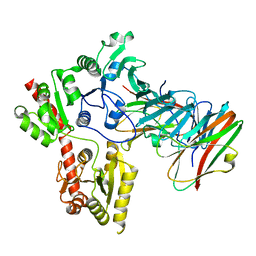 | | Crystal structure of accessory secretory protein 1,2 and 3 in Streptococcus pneumoniae | | Descriptor: | Accessory secretory protein Asp1, Accessory secretory protein Asp2, Accessory secretory protein Asp3 | | Authors: | Guo, C, Feng, Z, Zuo, G. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into the Asp1/2/3 complex mediated secretion of pneumococcal serine-rich repeat protein PsrP.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
5DRV
 
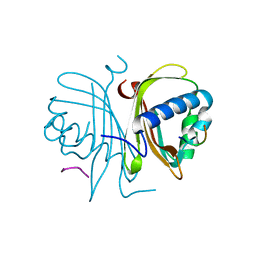 | |
8UUF
 
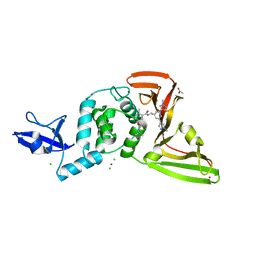 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun11941 | | Descriptor: | ACETATE ION, CHLORIDE ION, N-{(1R)-1-[(3M,5P)-3,5-bis(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-5-[2-(dimethylamino)ethoxy]-2-methylbenzamide, ... | | Authors: | Ansari, A, Tan, B, Riuz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UOB
 
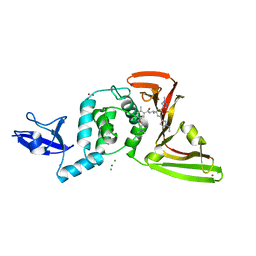 | | SARS-CoV-2 Papain-like protease (PLpro) with Inhibitor Jun12682 | | Descriptor: | 5-[2-(dimethylamino)ethoxy]-N-{(1R)-1-[(3M,5P)-3-(1-ethyl-1H-pyrazol-3-yl)-5-(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-2-methylbenzamide, CHLORIDE ION, Papain-like protease nsp3, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Wang, J, Arnold, E. | | Deposit date: | 2023-10-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
7CMD
 
 | | Crystal structure of the SARS-CoV-2 PLpro with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease.
Acta Pharm Sin B, 11, 2021
|
|
7CJD
 
 | |
5XWF
 
 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (SP3221/SAD) | | Descriptor: | Fungal chitinase from Rhizomucor miehei (SeMet-substituted proteins) | | Authors: | Jiang, Z.Q, Hu, S.Q, Liu, Y.C, Qin, Z, Yan, Q.J, Yang, S.Q. | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-04 | | Last modified: | 2021-08-18 | | Method: | X-RAY DIFFRACTION (2.581 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 2021
|
|
6WX4
 
 | |
6WUU
 
 | |
6W4Q
 
 | | Crystal structure of full-length tailspike protein 2 (TSP2, ORF211) ) from Escherichia coli O157:H7 bacteriophage CAB120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J, Herzberg, O. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7.
Sci Rep, 10, 2020
|
|
7PUR
 
 | | mouse Interleukin-12 subunit beta - p80 homodimer in space group P21 crystal form 2 | | Descriptor: | Interleukin-12 subunit beta, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2021-09-30 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Around she goes: the structure of mouse Interleukin-12 p80
To Be Published
|
|
