2ZDV
 
 | | Exploring Thrombin S1 pocket | | Descriptor: | D-phenylalanyl-N-(3-fluorobenzyl)-L-prolinamide, Hirudin variant-1, PHOSPHATE ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2007-11-29 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Exploring Thrombin S1 pocket
To be Published
|
|
7JYD
 
 | | Human Liver Receptor Homolog-1 in Complex with 10CA and a Fragment of Tif2 | | Descriptor: | 10-[(3aR,6R,6aR)-6-hydroxy-3-phenyl-3a-(1-phenylethenyl)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | D'Agostino, E.H, Mays, S.G, Ortlund, E.A. | | Deposit date: | 2020-08-30 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tapping into a phospholipid-LRH-1 axis yields a powerful anti-inflammatory agent with in vivo activity against colitis
To Be Published
|
|
7JYE
 
 | | Human Liver Receptor Homolog-1 in Complex with 9ChoP and a Fragment of Tif2 | | Descriptor: | 9-[(3~{a}~{R},6~{R},6~{a}~{R})-6-oxidanyl-3-phenyl-3~{a}-(1-phenylethenyl)-4,5,6,6~{a}-tetrahydro-1~{H}-pentalen-2-yl]nonyl 2-(trimethyl-$l^{4}-azanyl)ethyl hydrogen phosphate, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | D'Agostino, E.H, Mays, S.G, Ortlund, E.A. | | Deposit date: | 2020-08-30 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Tapping into a phospholipid-LRH-1 axis yields a powerful anti-inflammatory agent with in vivo activity against colitis
To Be Published
|
|
3HTV
 
 | |
1XBH
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(L-262) | | Descriptor: | PROTEIN (CYCLO(L-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-02-17 | | Release date: | 1999-02-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
1W2C
 
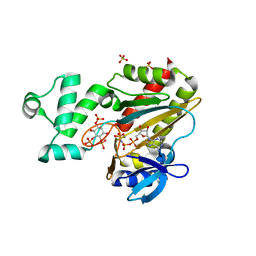 | | Human Inositol (1,4,5) trisphosphate 3-kinase complexed with Mn2+/AMPPNP/Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, MANGANESE (II) ION, ... | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
2ZDA
 
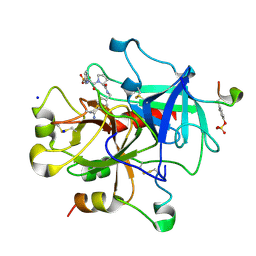 | | Exploring Thrombin S1 pocket | | Descriptor: | D-phenylalanyl-N-{4-[amino(iminio)methyl]benzyl}-L-prolinamide, Hirudin variant-1, PHOSPHATE ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2007-11-21 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
4KIR
 
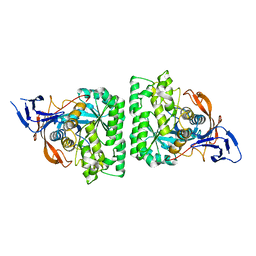 | |
2ZFF
 
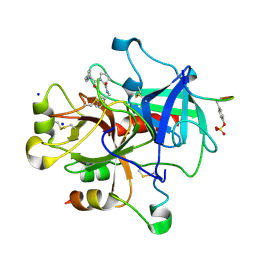 | | Exploring Thrombin S1-pocket | | Descriptor: | D-phenylalanyl-N-benzyl-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-04 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Exploring Thrombin S1-pocket
To be Published
|
|
8WLB
 
 | |
6O4M
 
 | | Racemic melittin | | Descriptor: | D-Melittin, Melittin, SULFATE ION | | Authors: | Kurgan, K.W, Bingman, C.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Retention of Native Quaternary Structure in Racemic Melittin Crystals.
J.Am.Chem.Soc., 141, 2019
|
|
8AHU
 
 | | Crystal structure of D-amino acid aminotrensferase from Haliscomenobacter hydrossis complexed with D-cycloserine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Mechanism of D-Cycloserine Inhibition of D-Amino Acid Transaminase from Haliscomenobacter hydrossis.
Biochemistry Mosc., 88, 2023
|
|
7E9W
 
 | | The Crystal Structure of D-psicose-3-epimerase from Biortus. | | Descriptor: | D-psicose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Wang, F, Xu, C, Qi, J, Zhang, M, Tian, F, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of D-psicose-3-epimerase from Biortus.
To Be Published
|
|
2ZC9
 
 | | Thrombin in complex with Inhibitor | | Descriptor: | D-phenylalanyl-N-(3-chlorobenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2007-11-06 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
2ZGB
 
 | | Thrombin Inhibition | | Descriptor: | D-leucyl-N-(3-chlorobenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Non-additivity of functional group contributions in protein-ligand binding: a comprehensive study by crystallography and isothermal titration calorimetry.
J.Mol.Biol., 397, 2010
|
|
6C0Y
 
 | |
2ZF0
 
 | | Exploring Thrombin S1 Pocket | | Descriptor: | D-phenylalanyl-N-(3-methylbenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2007-12-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring Thrombin S1 pocket
To be Published
|
|
2ZNK
 
 | | Thrombin Inhibition | | Descriptor: | D-leucyl-N-(4-carbamimidoylbenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-04-26 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-additivity of functional group contributions in protein-ligand binding: a comprehensive study by crystallography and isothermal titration calorimetry.
J.Mol.Biol., 397, 2010
|
|
4KQN
 
 | |
4R8J
 
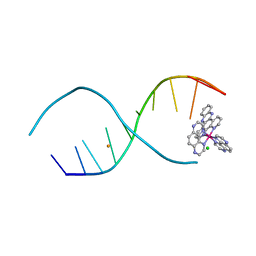 | | d(TCGGCGCCGA) with lambda-[Ru(TAP)2(dppz)]2+ soaked in D2O | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*(THM)P*CP*GP*GP*CP*GP*CP*CP*GP*A)-3'), ... | | Authors: | Hall, J.P, Gurung, S.P, Winter, G.W, Cardin, C.J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Monitoring one-electron photo-oxidation of guanine in DNA crystals using ultrafast infrared spectroscopy.
Nat Chem, 7, 2015
|
|
4IJC
 
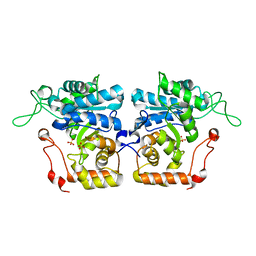 | | Crystal structure of arabinose dehydrogenase Ara1 from Saccharomyces cerevisiae | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, GLYCEROL, SULFATE ION | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-21 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IJR
 
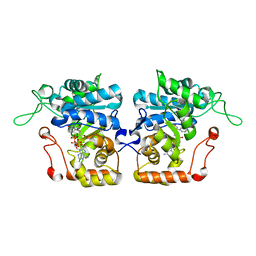 | | Crystal structure of Saccharomyces cerevisiae arabinose dehydrogenase Ara1 complexed with NADPH | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-23 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5FSR
 
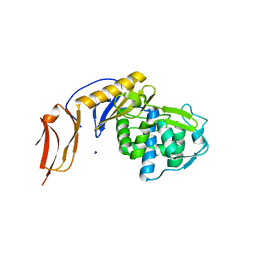 | | Crystal structure of penicillin binding protein 6B from Escherichia coli | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE DACD | | Authors: | Peters, K, Kannan, S, Rao, V.A, Bilboy, J, Vollmer, D, Erickson, S.W, Lewis, R.J, Young, K.D, Vollmer, W. | | Deposit date: | 2016-01-07 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Redundancy of Peptidoglycan Carboxypeptidases Ensures Robust Cell Shape Maintenance in Escherichia Coli
Mbio, 7, 2016
|
|
3GWQ
 
 | |
5MI1
 
 | | The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 9-th structure of the series with total exposition time 243 min. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2016-11-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural study of the X-ray-induced enzymatic reduction of molecular oxygen to water by Steccherinum murashkinskyi laccase: insights into the reaction mechanism.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
