1R16
 
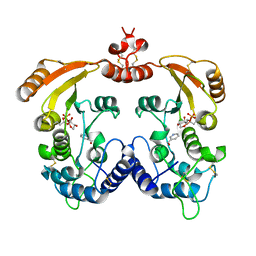 | | Aplysia ADP ribosyl cyclase with bound pyridylcarbinol and R5P | | Descriptor: | 3-PYRIDINYLCARBINOL, ADP-ribosyl cyclase, ANY 5'-MONOPHOSPHATE NUCLEOTIDE | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-23 | | Release date: | 2004-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
3IVF
 
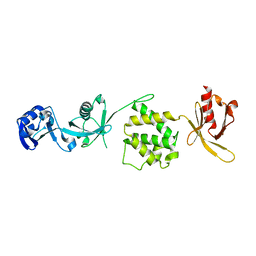 | | Crystal structure of the talin head FERM domain | | Descriptor: | Talin-1 | | Authors: | Elliott, P.R, Goult, B.T, Bate, N, Grossmann, J.G, Roberts, G.C.K, Critchley, D.R, Barsukov, I.L. | | Deposit date: | 2009-09-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Structure of the talin head reveals a novel extended conformation of the FERM domain
Structure, 18, 2010
|
|
1R55
 
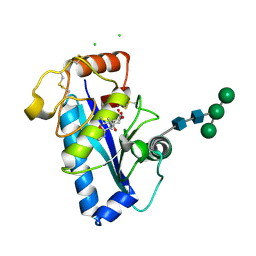 | | Crystal structure of the catalytic domain of human ADAM 33 | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAM 33, CALCIUM ION, ... | | Authors: | Orth, P, Reichert, P, Wang, W, Prosise, W.W, Yarosh-Tomaine, T, Hammond, G, Xiao, L, Mirza, U.A, Zou, J, Strickland, C, Taremi, S.S, Le, H.V, Madison, V. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structre of the catalytic domain of human ADAM33
J.Mol.Biol., 335, 2004
|
|
5IVV
 
 | |
5IWG
 
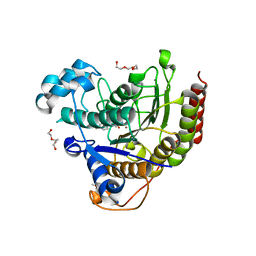 | | HDAC2 WITH LIGAND BRD4884 | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Steinbacher, S. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Kinetic and structural insights into the binding of histone deacetylase 1 and 2 (HDAC1, 2) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
3IPU
 
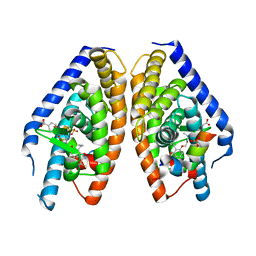 | | X-ray structure of benzisoxazole urea synthetic agonist bound to the LXR-alpha | | Descriptor: | 4-{[methyl(3-{[7-propyl-3-(trifluoromethyl)-1,2-benzisoxazol-6-yl]oxy}propyl)carbamoyl]amino}benzoic acid, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
5J7T
 
 | |
1BSE
 
 | |
1RBP
 
 | |
5J0O
 
 | |
5J0W
 
 | |
4XOI
 
 | | Structure of hsAnillin bound with RhoA(Q63L) at 2.1 Angstroms resolution | | Descriptor: | Actin-binding protein anillin, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sun, L, Guan, R, Lee, I.-J, Liu, Y, Chen, M, Wang, J, Wu, J, Chen, Z. | | Deposit date: | 2015-01-16 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Mechanistic insights into the anchorage of the contractile ring by anillin and mid1
Dev.Cell, 33, 2015
|
|
4XOZ
 
 | | Crystal structure of ERK2 in complex with an inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, N~1~-[3-(benzyloxy)benzyl]-1H-tetrazole-1,5-diamine, SULFATE ION | | Authors: | Gelin, M, Allemand, F, Labesse, G, Guichou, J.F. | | Deposit date: | 2015-01-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5ITT
 
 | | Crystal Structure of Human NEIL1 bound to duplex DNA containing THF | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1, GLYCEROL | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3I85
 
 | |
1RKH
 
 | | crystal structure of the rat vitamin D receptor ligand binding domain complexed with 2AM20R and a synthetic peptide containing the NR2 box of DRIP 205 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-2-METHYL-CYCLOHEXANE-1,3-DIOL, Peroxisome proliferator-activated receptor binding protein, Vitamin D3 receptor | | Authors: | Vanhooke, J.L, Benning, M.M, Bauer, C.B, Pike, J.W, F DeLuca, H. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular Structure of the Rat Vitamin D Receptor Ligand Binding Domain Complexed with 2-Carbon-Substituted Vitamin D(3) Hormone Analogues and a LXXLL-Containing Coactivator Peptide
Biochemistry, 43, 2004
|
|
5ITU
 
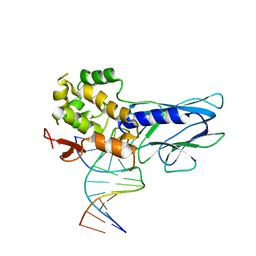 | | Crystal Structure of Human NEIL1(242K) bound to duplex DNA containing THF | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*CP*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1BU8
 
 | | RAT PANCREATIC LIPASE RELATED PROTEIN 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (PANCREATIC LIPASE RELATED PROTEIN 2) | | Authors: | Roussel, A, Cambillau, C. | | Deposit date: | 1998-09-14 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of rat pancreatic lipase-related protein 2.
J.Biol.Chem., 273, 1998
|
|
4XBF
 
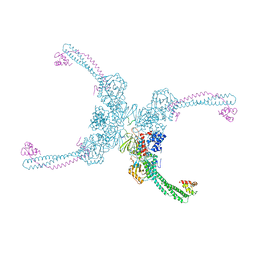 | | Structure of LSD1:CoREST in complex with ssRNA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Luka, Z, Loukachevitch, L.V, Martin, W.J, Wagner, C, Reiter, N.J. | | Deposit date: | 2014-12-16 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | G-quadruplex RNA binding and recognition by the lysine-specific histone demethylase-1 enzyme.
RNA, 22, 2016
|
|
5IX0
 
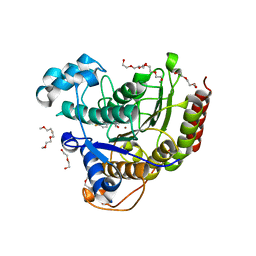 | | HDAC2 WITH LIGAND BRD7232 | | Descriptor: | (3-exo)-N-(4-amino-4'-fluoro[1,1'-biphenyl]-3-yl)-8-oxabicyclo[3.2.1]octane-3-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Steinbacher, S. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic and structural insights into the binding of histone deacetylase 1 and 2 (HDAC1, 2) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5TO8
 
 | | Selectivity switch between FAK and Pyk2: Macrocyclization of FAK inhibitors improves Pyk2 potency | | Descriptor: | 25-(methylsulfonyl)-8-(trifluoromethyl)-5,17,18,21,22,23,24,25-octahydro-12H-7,11-(azeno)-16,13-(metheno)pyrido[3,2-i]pyrrolo[1,2-q][1,3,7,11,17]pentaazacyclohenicosin-20(6H)-one, Protein-tyrosine kinase 2-beta | | Authors: | Newby, Z.E. | | Deposit date: | 2016-10-17 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9849 Å) | | Cite: | Selectivity switch between FAK and Pyk2: Macrocyclization of FAK inhibitors improves Pyk2 potency.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
3IDO
 
 | |
1C3K
 
 | | CRYSTAL STRUCTURE OF HELIANTHUS TUBEROSUS LECTIN | | Descriptor: | AGGLUTININ | | Authors: | Bourne, Y, Zamboni, V, Barre, A, Peumans, W.J, van Damme, E.J.M, Rouge, P. | | Deposit date: | 1999-07-28 | | Release date: | 2000-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helianthus tuberosus lectin reveals a widespread scaffold for mannose-binding lectins.
Structure Fold.Des., 7, 1999
|
|
3II0
 
 | | Crystal structure of human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase, cytoplasmic, ... | | Authors: | Ugochukwu, E, Pilka, E, Cooper, C, Bray, J.E, Yue, W.W, Muniz, J, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human Glutamate oxaloacetate transaminase 1 (GOT1)
To be Published
|
|
4XZP
 
 | |
