8U5R
 
 | |
8U5F
 
 | | Crystal Structure of Trypsinized Clostridium perfringens Enterotoxin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Heat-labile enterotoxin B chain, ... | | Authors: | Kapoor, S, Ogbu, C.P, Vecchio, A.J. | | Deposit date: | 2023-09-12 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin.
Toxins, 15, 2023
|
|
8U5E
 
 | |
8U5D
 
 | |
8U5B
 
 | |
8U51
 
 | |
8U50
 
 | |
8U4Z
 
 | |
8U4V
 
 | |
8U4T
 
 | |
8U4S
 
 | |
8U4F
 
 | |
8U4A
 
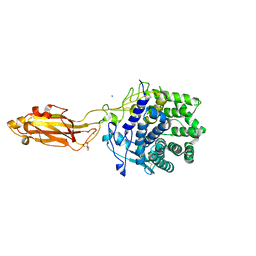 | |
8U49
 
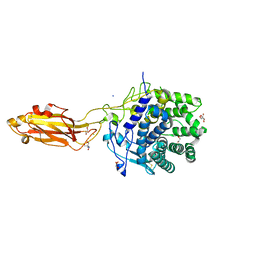 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
8U3Z
 
 | | Model of TTLL6 bound to microtubule from composite map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-09-08 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 2024
|
|
8U3K
 
 | | DdmDE handover complex | | Descriptor: | DNA (29-MER), DNA (5'-D(P*GP*GP*AP*AP*AP*TP*GP*TP*TP*GP*AP*AP*TP*AP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Bravo, J.P.K. | | Deposit date: | 2023-09-07 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
8U3B
 
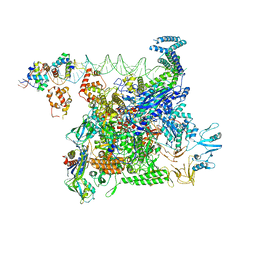 | | Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A | | Descriptor: | DNA (69-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Kompaniiets, D, Wang, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for transcription activation by the nitrate-responsive regulator NarL.
Nucleic Acids Res., 52, 2024
|
|
8U37
 
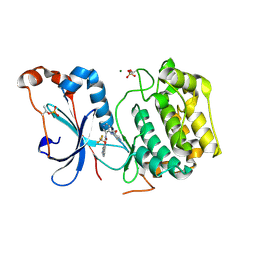 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with NVP-CJL037 at 2.48-A resolution | | Descriptor: | (6M)-3-amino-N-{4-[(3R,4S)-4-amino-3-methoxypiperidin-1-yl]pyridin-3-yl}-6-[3-(trifluoromethoxy)pyridin-2-yl]pyrazine-2-carboxamide, MAGNESIUM ION, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
8U31
 
 | | Crystal structure of PD-1 in complex with a Fab | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Sun, D, Masureel, M. | | Deposit date: | 2023-09-07 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure- and machine learning-guided engineering demonstrate that a non-canonical disulfide in an anti-PD-1 rabbit antibody does not impede antibody developability.
Mabs, 16, 2024
|
|
8U2X
 
 | |
8U2U
 
 | |
8U2S
 
 | |
8U2O
 
 | |
8U2E
 
 | | Bruton's tyrosine kinase in complex with N-[(2R)-1-[(3R)-3-(methylcarbamoyl)-1H,2H,3H,4H,9H-pyrido[3,4-b]indol-2-yl]-3-(3-methylphenyl)-1-oxopropan-2-yl]-1H-indazole-5-carboxamide | | Descriptor: | (2S)-6-fluoro-5-[(3S)-3-(3-methyl-2-oxoimidazolidin-1-yl)piperidin-1-yl]-2-(4-methylpiperazine-1-carbonyl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Gajewski, S, Clifton, M.C. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Preclinical Pharmacology of NX-2127, an Orally Bioavailable Degrader of Bruton's Tyrosine Kinase with Immunomodulatory Activity for the Treatment of Patients with B Cell Malignancies.
J.Med.Chem., 67, 2024
|
|
8U2D
 
 | | Bruton's tyrosine kinase in complex with N-[(2R)-1-[(3R)-3-(methylcarbamoyl)-1H,2H,3H,4H,9H-pyrido[3,4-b]indol-2-yl]-3-(3-methylphenyl)-1-oxopropan-2-yl]-1H-indazole-5-carboxamide | | Descriptor: | (3R)-2-[N-(1H-indazole-5-carbonyl)-3-methyl-D-phenylalanyl]-N-methyl-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Gajewski, S, Clifton, M.C. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Preclinical Pharmacology of NX-2127, an Orally Bioavailable Degrader of Bruton's Tyrosine Kinase with Immunomodulatory Activity for the Treatment of Patients with B Cell Malignancies.
J.Med.Chem., 67, 2024
|
|
