3QUI
 
 | | Crystal structure of Pseudomonas aeruginosa Hfq in complex with ADPNP | | Descriptor: | ADENINE, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Murina, V.N, Gabdulkhakov, A.G. | | Deposit date: | 2011-02-24 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Hfq binds ribonucleotides in three different RNA-binding sites.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8OXU
 
 | | Crystal Structure of the Hsp90-LA1011 Complex | | Descriptor: | ATP-dependent molecular chaperone HSP82, dimethyl 2,6-bis[2-(dimethylamino)ethyl]-1-methyl-4-[4-(trifluoromethyl)phenyl]-4~{H}-pyridine-3,5-dicarboxylate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2023-05-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer's Disease.
Biomolecules, 13, 2023
|
|
8P01
 
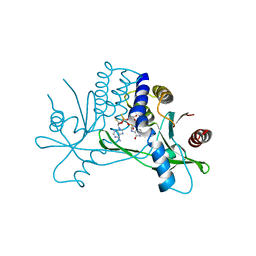 | | Crystal structure of human STING ectodomain in complex with BI 7446, a potent cyclic dinucleotide STING agonist with broad-spectrum variant activity for the treatment of cancer | | Descriptor: | 3-[(1~{R},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9-fluoranyl-18-oxidanyl-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-6~{H}-imidazo[4,5-d]pyridazin-7-one, Stimulator of interferon genes protein | | Authors: | Nar, H. | | Deposit date: | 2023-05-09 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Discovery of BI 7446: A Potent Cyclic Dinucleotide STING Agonist with Broad-Spectrum Variant Activity for the Treatment of Cancer.
J.Med.Chem., 66, 2023
|
|
1YIQ
 
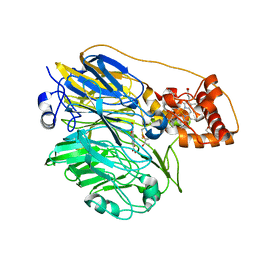 | | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADHIIG from Pseudomonas putida HK5. Compariison to the other quinohemoprotein alcohol dehydrogenase ADHIIB found in the same microorganism. | | Descriptor: | CALCIUM ION, HEME C, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Toyama, H, Chen, Z.W, Fukumoto, M, Adachi, O, Matsushita, K, Mathews, F.S. | | Deposit date: | 2005-01-12 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADH-IIG from Pseudomonas putida HK5
J.Mol.Biol., 352, 2005
|
|
8OLU
 
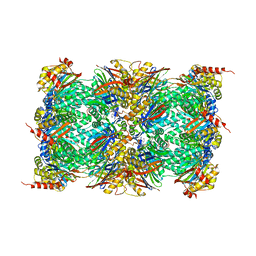 | |
3RAP
 
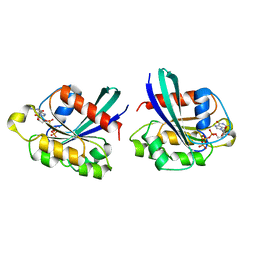 | |
1OUB
 
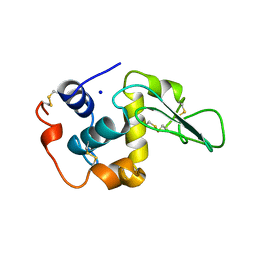 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V100A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1OUH
 
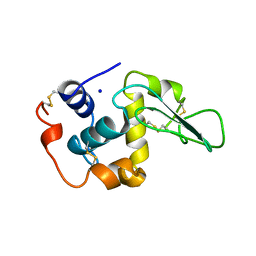 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V74A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
1OUF
 
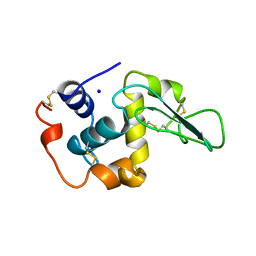 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V130A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
3SKB
 
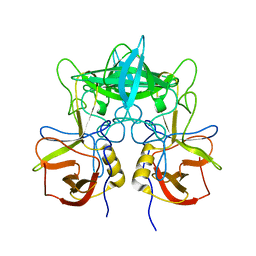 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) | | Descriptor: | Capsid | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
8PKY
 
 | |
8PM1
 
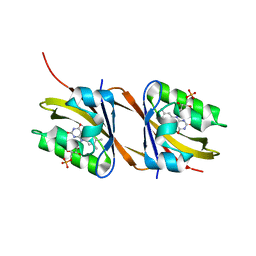 | |
2Q09
 
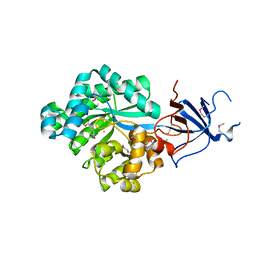 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | Descriptor: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
3TMN
 
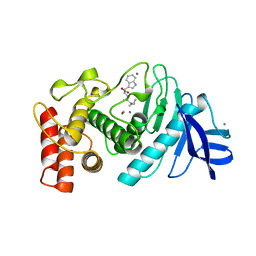 | |
3TQX
 
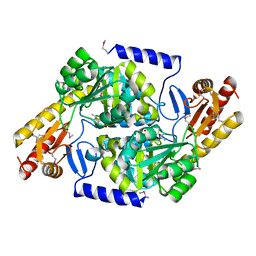 | | Structure of the 2-amino-3-ketobutyrate coenzyme A ligase (kbl) from Coxiella burnetii | | Descriptor: | 2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
8PEK
 
 | |
1ZNN
 
 | | Structure of the synthase subunit of PLP synthase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PLP SYNTHASE, SULFATE ION | | Authors: | Zhu, J, Burgner, J.W, Harms, E, Belitsky, B.R, Smith, J.L. | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A New Arrangement of (beta/alpha)8 Barrels in the Synthase Subunit of PLP Synthase.
J.Biol.Chem., 280, 2005
|
|
2FLI
 
 | | The crystal structure of D-ribulose 5-phosphate 3-epimerase from Streptococus pyogenes complexed with D-xylitol 5-phosphate | | Descriptor: | D-XYLITOL-5-PHOSPHATE, ZINC ION, ribulose-phosphate 3-epimerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Akana, J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | d-Ribulose 5-Phosphate 3-Epimerase: Functional and Structural Relationships to Members of the Ribulose-Phosphate Binding (beta/alpha)(8)-Barrel Superfamily(,).
Biochemistry, 45, 2006
|
|
3R05
 
 | |
3SEJ
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to Secretor Lewis HBGA (LeB) | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
1XQZ
 
 | | Crystal Structure of hPim-1 kinase at 2.1 A resolution | | Descriptor: | Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Qian, K.C, Wang, L, Hickey, E.R, Studts, J, Barringer, K, Peng, C, Kronkaitis, A, Li, J, White, A, Mische, S, Farmer, B. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Constitutive Activity and a Unique Nucleotide Binding Mode of Human Pim-1 Kinase.
J.Biol.Chem., 280, 2005
|
|
3SLD
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to A trisaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
8PX2
 
 | |
3S48
 
 | | Human Alpha-Haemoglobin Complexed with the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Hemoglobin subunit alpha, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kumar, K.K, Jacques, D.A, Caradoc-Davies, T.T, Guss, J.M, Gell, D.A. | | Deposit date: | 2011-05-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The structure of alpha-haemoglobin in complex with a haemoglobin-binding domain from Staphylococcus aureus reveals the elusive alpha-haemoglobin dimerization interface
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
8PXA
 
 | |
