4D36
 
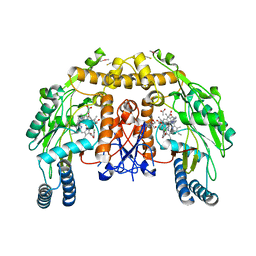 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with N-2-(2-(1H-imidazol-1-yl)pyrimidin-4-yl)ethyl-3-(3- chlorophenyl)propan-1-amine | | Descriptor: | 3-(3-chlorophenyl)-N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}propan-1-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
4D88
 
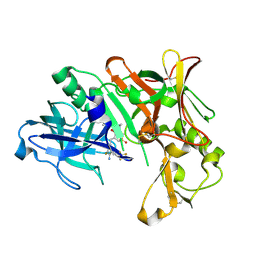 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BXQ490 | | Descriptor: | (3S,4S,5R)-3-{4-amino-3-fluoro-5-[(2S)-3,3,3-trifluoro-2-hydroxypropyl]benzyl}-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
3NU6
 
 | |
4D85
 
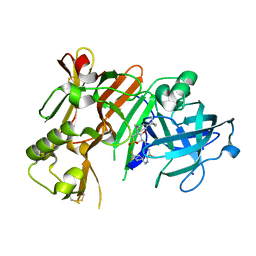 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BVI151 | | Descriptor: | (3R,4S,5S)-3-[(3-tert-butylbenzyl)amino]-5-[(4,4,7'-trifluoro-1',2'-dihydrospiro[cyclohexane-1,3'-indol]-5'-yl)methyl]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1, IODIDE ION | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
7QFE
 
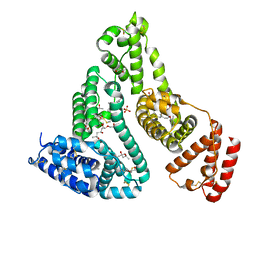 | | Crystal structure of Human Serum albumin in complex with Gemfibrozil | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(2,5-dimethylphenoxy)-2,2-dimethylpentanoic acid, MYRISTIC ACID, ... | | Authors: | Liberi, S, Moro, G, Angelini, A, Cendron, L. | | Deposit date: | 2021-12-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Human Serum Albumin in Complex with the Fibrate Drug Gemfibrozil.
Int J Mol Sci, 23, 2022
|
|
3NW2
 
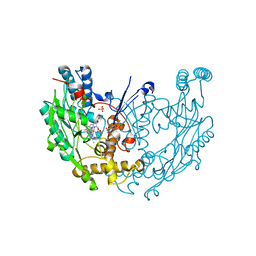 | | Novel nanomolar Imidazopyridines as selective Nitric Oxide Synthase (iNOS) inhibitors: SAR and structural insights | | Descriptor: | 2-[2-(4-methoxypyridin-2-yl)ethyl]-3H-imidazo[4,5-b]pyridine, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Graedler, U, Fuchss, T, Ulrich, W.R, Boer, R, Strub, A, Hesslinger, C, Anezo, C, Diederichs, K, Zaliani, A. | | Deposit date: | 2010-07-09 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel nanomolar imidazo[4,5-b]pyridines as selective nitric oxide synthase (iNOS) inhibitors: SAR and structural insights
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4D2Y
 
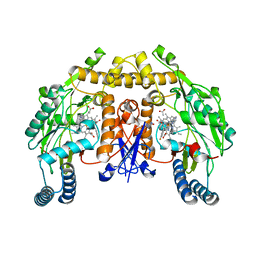 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (1R,2R)-2-(3-fluorobenzyl)-N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}cyclopropanamine | | Descriptor: | (1R,2R)-2-(3-fluorobenzyl)-N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}cyclopropanamine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
7ZRP
 
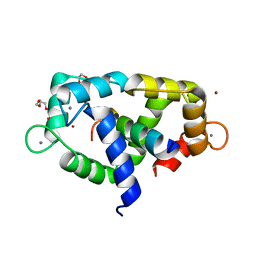 | | 2.65 Angstrom crystal structure of Ca/CaM:CaMKIIdelta peptide complex | | Descriptor: | CALCIUM ION, Calcium/calmodulin-dependent protein kinase type II subunit delta, Calmodulin-1, ... | | Authors: | Helassa, N, Antonyuk, S. | | Deposit date: | 2022-05-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Calmodulin variant E140G associated with long QT syndrome impairs CaMKII delta autophosphorylation and L-type calcium channel inactivation.
J.Biol.Chem., 299, 2022
|
|
4QB3
 
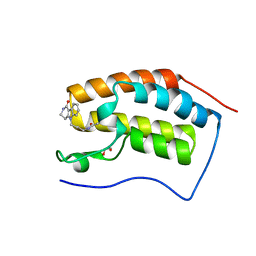 | | Crystal structure of the first bromodomain of human BRD4 in complex with Olinone | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-[4-(1-oxo-1,2,3,4-tetrahydro-5H-pyrido[4,3-b]indol-5-yl)butyl]acetamide | | Authors: | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | Deposit date: | 2014-05-06 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression.
Chem.Biol., 21, 2014
|
|
4Q7F
 
 | | 1.98 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with Adenosine. | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, 5' nucleotidase family protein, MAGNESIUM ION, ... | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1.98 Angstrom Crystal Structure of Putative 5'-Nucleotidase from Staphylococcus aureus in complex with Adenosine.
TO BE PUBLISHED
|
|
4DJH
 
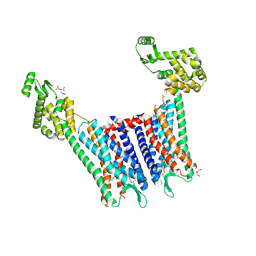 | | Structure of the human kappa opioid receptor in complex with JDTic | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (3R)-7-hydroxy-N-{(2S)-1-[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]-3-methylbutan-2-yl}-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, CITRIC ACID, ... | | Authors: | Wu, H, Wacker, D, Katritch, V, Mileni, M, Han, G.W, Vardy, E, Liu, W, Thompson, A.A, Huang, X.P, Carroll, F.I, Mascarella, S.W, Westkaemper, R.B, Mosier, P.D, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-02-01 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | tructure of the human kappa-opioid receptor in complex with JDTic
Nature, 485, 2012
|
|
5UEN
 
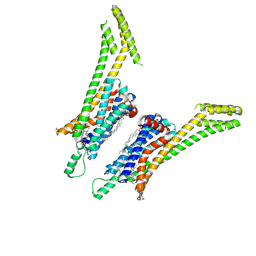 | | Crystal structure of the human adenosine A1 receptor A1AR-bRIL in complex with the covalent antagonist DU172 at 3.2A resolution | | Descriptor: | 4-{[3-(8-cyclohexyl-2,6-dioxo-1-propyl-1,2,6,7-tetrahydro-3H-purin-3-yl)propyl]carbamoyl}benzene-1-sulfonyl fluoride, Adenosine receptor A1,Soluble cytochrome b562,Adenosine receptor A1, OLEIC ACID | | Authors: | Glukhova, A, Thal, D.M, Nguyen, A.T, Vecchio, E.A, Jorg, M, Scammells, P.J, May, L.T, Sexton, P.M, Christopoulos, A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity.
Cell, 168, 2017
|
|
4DPF
 
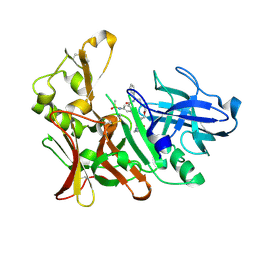 | | BACE-1 in complex with a HEA-macrocyclic type inhibitor | | Descriptor: | Beta-secretase 1, N-[(4S,8E,11S)-4-[(1R)-1-hydroxy-2-{[3-(propan-2-yl)benzyl]amino}ethyl]-2,13-dioxo-11-phenyl-6-oxa-3,12-diazabicyclo[12.3.1]octadeca-1(18),8,14,16-tetraen-16-yl]-N-methylmethanesulfonamide | | Authors: | Lindberg, J, Borkakoti, N, Derbyshire, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-07-11 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly potent macrocyclic BACE-1 inhibitors incorporating a hydroxyethylamine core: design, synthesis and X-ray crystal structures of enzyme inhibitor complexes.
Bioorg.Med.Chem., 20, 2012
|
|
4DC3
 
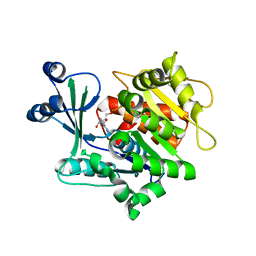 | | Adenosine kinase from Schistosoma mansoni in complex with 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, ADENOSINE, Adenosine kinase, ... | | Authors: | Romanello, L, Bachega, F.R, Garatt, R.C, DeMarco, R, Brandao-neto, J, Pereira, H.M. | | Deposit date: | 2012-01-17 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Adenosine kinase from Schistosoma mansoni: structural basis for the differential incorporation of nucleoside analogues.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MM5
 
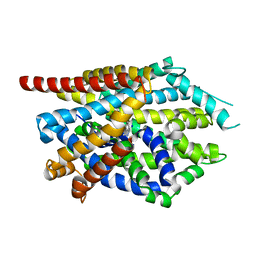 | | Crystal structure of LeuBAT (delta13 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
4MMB
 
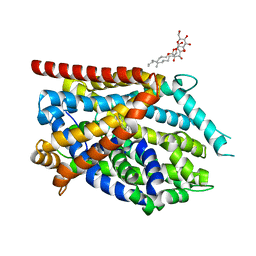 | | Crystal structure of LeuBAT (delta6 mutant) in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, SODIUM ION, Transporter, ... | | Authors: | Wang, H, Gouaux, E. | | Deposit date: | 2013-09-08 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for action by diverse antidepressants on biogenic amine transporters.
Nature, 503, 2013
|
|
5U6C
 
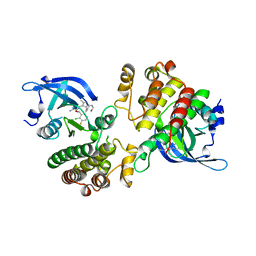 | | Crystal structure of the Mer kinase domain in complex with a macrocyclic inhibitor | | Descriptor: | (10R)-7-amino-11-chloro-12-fluoro-1-(2-hydroxyethyl)-3,10,16-trimethyl-16,17-dihydro-1H-8,4-(azeno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, Tyrosine-protein kinase Mer | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Axl kinase domain in complex with a macrocyclic inhibitor offers first structural insights into an active TAM receptor kinase.
J. Biol. Chem., 292, 2017
|
|
7ZMY
 
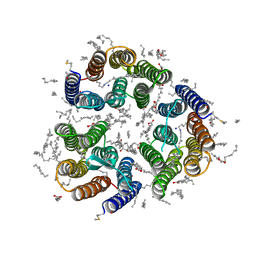 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN0
 
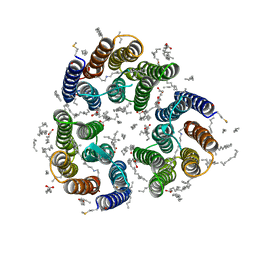 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN3
 
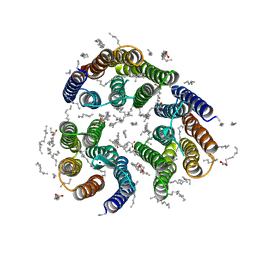 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the L state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN9
 
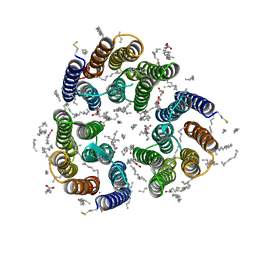 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 7.0 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNB
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 5.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNA
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 5.2 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZND
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 7.6 in the absence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN8
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 7.0 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
