1KMX
 
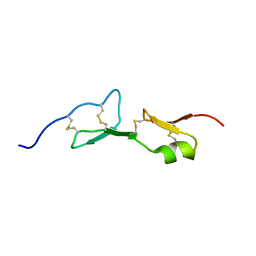 | |
1HRA
 
 | | THE SOLUTION STRUCTURE OF THE HUMAN RETINOIC ACID RECEPTOR-BETA DNA-BINDING DOMAIN | | Descriptor: | RETINOIC ACID RECEPTOR, ZINC ION | | Authors: | Knegtel, R.M.A, Katahira, M, Schilthuis, J.G, Bonvin, A.M.J.J, Boelens, R, Eib, D, Van Der Saag, P.T, Kaptein, R. | | Deposit date: | 1993-07-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the human retinoic acid receptor-beta DNA-binding domain.
J.Biomol.NMR, 3, 1993
|
|
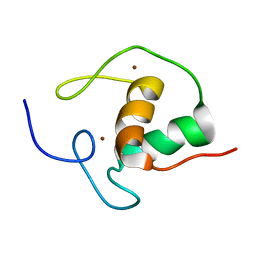
1E0E
 
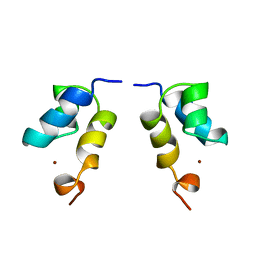 | | N-terminal zinc-binding HHCC domain of HIV-2 integrase | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS TYPE 2 INTEGRASE, ZINC ION | | Authors: | Eijkelenboom, A.P.A.M, Van Den ent, F.M.I, Plasterk, R.H.A, Kaptein, R, Boelens, R. | | Deposit date: | 2000-03-25 | | Release date: | 2001-03-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the Dimeric N-Terminal Hhcc Domain of HIV-2 Integrase
J.Biomol.NMR, 18, 2000
|
|
1QTU
 
 | | SOLUTION STRUCTURE OF THE ONCOPROTEIN P13MTCP1 | | Descriptor: | PROTEIN (PRODUCT OF THE MTCP1 ONCOGENE) | | Authors: | Guignard, L, Padilla, A, Mispelter, J, Yang, Y.-S, Stern, M.-H, Lhoste, J.-M, Roumestand, C. | | Deposit date: | 1999-06-29 | | Release date: | 2001-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and solution structure refinement of the 15N-labeled human oncogenic protein p13MTCP1: comparison with X-ray data.
J.Biomol.NMR, 17, 2000
|
|
1WQU
 
 | | Solution structure of the human FES SH2 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase FES/FPS | | Authors: | Scott, A, Pantoja-Uceda, D, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Tanaka, A, Sugano, S, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-02 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Src homology 2 domain from the human feline sarcoma oncogene Fes
J.Biomol.NMR, 31, 2005
|
|
2BJX
 
 | | PROTEIN DISULFIDE ISOMERASE | | Descriptor: | PROTEIN (PROTEIN DISULFIDE ISOMERASE) | | Authors: | Kemmink, J, Dijkstra, K, Mariani, M, Scheek, R.M, Penka, E, Nilges, M, Darby, N.J. | | Deposit date: | 1999-01-30 | | Release date: | 1999-02-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structure in solution of the b domain of protein disulfide isomerase.
J.Biomol.NMR, 13, 1999
|
|
2K4C
 
 | | tRNAPhe-based homology model for tRNAVal refined against base N-H RDCs in two media and SAXS data | | Descriptor: | 76-MER | | Authors: | Grishaev, A, Ying, J, Canny, M.D, Pardi, A, Bax, A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data.
J.Biomol.Nmr, 42, 2008
|
|
7OTD
 
 | | Oxytocin NMR solution structure | | Descriptor: | AMINO GROUP, COPPER (II) ION, UNK-TYR-ILE-GLN-ASN-CYS-PRO-LEU-GLY | | Authors: | Shalev, D.E, Alshanski, I, Yitzchaik, S, Hurevich, M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-13 | | Method: | SOLUTION NMR | | Cite: | Determining the structure and binding mechanism of oxytocin-Cu 2+ complex using paramagnetic relaxation enhancement NMR analysis.
J.Biol.Inorg.Chem., 26, 2021
|
|
7OFG
 
 | |
6TWE
 
 | |
6U3S
 
 | |
5OLF
 
 | |
6U6Q
 
 | |
4TGF
 
 | | SOLUTION STRUCTURES OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA DERIVED FROM 1*H NMR DATA | | Descriptor: | DES-VAL-1,VAL-2,TRANSFORMING GROWTH FACTOR, ALPHA | | Authors: | Kline, T.P, Brown, F.K, Brown, S.C, Jeffs, P.W, Kopple, K.D, Mueller, L. | | Deposit date: | 1990-06-13 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human transforming growth factor alpha derived from 1H NMR data.
Biochemistry, 29, 1990
|
|
1W6B
 
 | | Solution NMR Structure of a Long Neurotoxin from the Venom of the Asian Cobra, 20 Structures | | Descriptor: | LONG NEUROTOXIN 1 | | Authors: | Talebzadeh-Farooji, M, Amininasab, M, Elmi, M.M, Naderi-Manesh, H, Sarbolouki, M.N. | | Deposit date: | 2004-08-17 | | Release date: | 2004-12-22 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of long neurotoxin NTX-1 from the venom of Naja naja oxiana by 2D-NMR spectroscopy.
Eur. J. Biochem., 271, 2004
|
|
3MEF
 
 | |
6WA1
 
 | |
9CPI
 
 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-[1-(2-aminoethyl)-4,5-dihydro-1H-imidazol-2-yl]-N-[4-(4,5-dihydro-1H-imidazol-2-yl)phenyl]benzamide, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | NMR structures of small molecules bound to a model of a CUG RNA repeat expansion.
Bioorg.Med.Chem.Lett., 111, 2024
|
|
1M8M
 
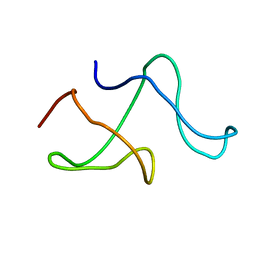 | | SOLID-STATE MAS NMR STRUCTURE OF THE A-SPECTRIN SH3 DOMAIN | | Descriptor: | SPECTRIN ALPHA CHAIN, BRAIN | | Authors: | Castellani, F, Van Rossum, B, Diehl, A, Schubert, M, Rehbein, K, Oschkinat, H. | | Deposit date: | 2002-07-25 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy
Nature, 420, 2002
|
|
8X4F
 
 | |
9CJA
 
 | |
8H1D
 
 | | Solid-state NMR Structure of Aquaporin Z in its Native Cellular Membranes | | Descriptor: | Aquaporin Z | | Authors: | Xie, H, Zhao, Y, Zhao, W, Chen, Y, Liu, M, Yang, J. | | Deposit date: | 2022-10-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR structure determination of a membrane protein in E. coli cellular inner membrane.
Sci Adv, 9, 2023
|
|
8GQO
 
 | |
5ODD
 
 | | HUMAN MED26 N-TERMINAL DOMAIN (1-92) | | Descriptor: | Mediator of RNA polymerase II transcription subunit 26 | | Authors: | Lens, Z, Cantrelle, F.-X, Perruzini, R, Dewitte, F, Hanoulle, X, Villeret, V, Verger, A, Landrieu, I. | | Deposit date: | 2017-07-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C assignments of the N-terminal domain of the Mediator complex subunit MED26.
Biomol.Nmr Assign., 10, 2016
|
|
5OPH
 
 | |
