3W87
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-103 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[5-(methoxycarbonyl)naphthalen-2-yl]butyl}-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, CACODYLATE ION, ... | | Authors: | Inaoka, D.K, Hashimoto, S, Rocha, J.R, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Kuranaga, T, Shiba, T, Balogun, E.O, Sakamoto, K, Suzuki, S, Montanari, C.A, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-103
To be Published
|
|
3W86
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-96 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[4-(methoxycarbonyl)phenyl]butyl}-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, CACODYLATE ION, ... | | Authors: | Inaoka, D.K, Hashimoto, S, Rocha, J.R, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Kuranaga, T, Shiba, T, Balogun, E.O, Sakamoto, K, Suzuki, S, Montanari, C.A, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-96
To be Published
|
|
7KBN
 
 | | 1.60 Angstrom resolution crystal structure of the beta-Q114A mutant of Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site, and 2-aminophenol quinonoid at the enzyme beta site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-10-02 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of the beta-Q114A mutant of Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site, and 2-aminophenol quinonoid at the enzyme beta site.
To be Published
|
|
4KTT
 
 | | Structural insights of MAT enzymes: MATa2b complexed with SAM | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine adenosyltransferase 2 subunit beta, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2013-05-21 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure and function study of the complex that synthesizes S-adenosylmethionine.
IUCrJ, 1, 2014
|
|
4OOE
 
 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203Y mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
3NUS
 
 | |
4EJI
 
 | |
1MG5
 
 | | Crystal structure of Drosophila melanogaster alcohol dehydrogenase complexed with NADH and acetate at 1.6 A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, alcohol dehydrogenase | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 2002-08-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Drosophila alcohol dehydrogenase: acetate-enzyme interactions and novel insights into the effects of electrostatics on catalysis
J.Mol.Biol., 345, 2005
|
|
4QJQ
 
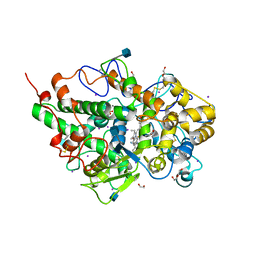 | | Crystal structure of goat lactoperoxidase in complex with octopamine at 2.1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, ... | | Authors: | Singh, R.P, Kushwaha, G.S, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-06-04 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of goat lactoperoxidase in complex with octopamine at 2.1 Angstrom resolution
To be Published
|
|
1MAR
 
 | |
3HKP
 
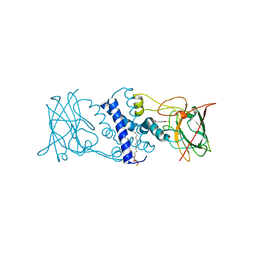 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with protocatechuate | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 3,4-DIHYDROXYBENZOIC ACID, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-25 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
4IFA
 
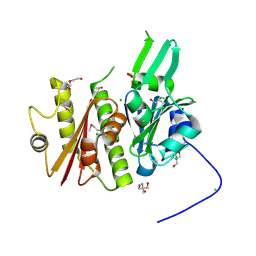 | | 1.5 Angstrom resolution crystal structure of an extracellular protein containing a SCP domain from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Shatsman, S, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-14 | | Release date: | 2012-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom resolution crystal structure of an extracellular protein containing a SCP domain from Bacillus anthracis str. Ames
To be Published
|
|
5QII
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-(3-(1-(4-CHLOROPHENYL)CYCLOPROPYL) -[1,2,4]TRIAZOLO[4,3-A]PYRIDIN-8-YL)PROPAN-2-OL | | Descriptor: | 2-{3-[1-(4-chlorophenyl)cyclopropyl][1,2,4]triazolo[4,3-a]pyridin-8-yl}propan-2-ol, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of Clinical Candidate BMS-823778 as an Inhibitor of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 9, 2018
|
|
4RGQ
 
 | | Crystal structure of the Methanocaldococcus jannaschii G1PDH with NADPH and DHAP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, Glycerol-1-phosphate dehydrogenase, ... | | Authors: | Carbone, V, Ronimus, R.S, Schofield, L.R, Sutherland-Smith, A.J. | | Deposit date: | 2014-09-30 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure and Evolution of the Archaeal Lipid Synthesis Enzyme sn-Glycerol-1-phosphate Dehydrogenase.
J.Biol.Chem., 290, 2015
|
|
4CLO
 
 | |
5ONI
 
 | | LOW-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,4-BUTANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
5U6V
 
 | | X-ray crystal structure of 1,2,3-triazolobenzodiazepine in complex with BRD2(D2) | | Descriptor: | 1,2-ETHANEDIOL, 5-[7-(4-chlorophenyl)-1-methyl-6,7-dihydro-5H-[1,2,3]triazolo[1,5-d][1,4]benzodiazepin-9-yl]pyridin-2-amine, Bromodomain-containing protein 2 | | Authors: | Hatfaludi, T, Sharp, P.P, Garnier, J.-M, Burns, C.J, Czabotar, P.E. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Design, Synthesis, and Biological Activity of 1,2,3-Triazolobenzodiazepine BET Bromodomain Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5QIJ
 
 | | CRYSTAL STRUCTURE OF MURINE 11B- HYDROXYSTEROIDDEHYDROGENASE COMPLEXED WITH 2-(3-(1-(4- CHLOROPHENYL)CYCLOPROPYL)-[1,2,4]TRIAZOLO[4,3-A]PYRIDIN-8- YL)PROPAN-2-OL | | Descriptor: | 2-{3-[1-(4-chlorophenyl)cyclopropyl][1,2,4]triazolo[4,3-a]pyridin-8-yl}propan-2-ol, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of Clinical Candidate BMS-823778 as an Inhibitor of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 9, 2018
|
|
3LFL
 
 | | Crystal Structure of human Glutathione Transferase Omega 1, delta 155 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLUTATHIONE, Glutathione S-transferase omega-1 | | Authors: | Brock, J. | | Deposit date: | 2010-01-18 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel folding and stability defects cause a deficiency of human glutathione transferase omega 1.
J.Biol.Chem., 286, 2011
|
|
2WGZ
 
 | | Crystal structure of alpha-1,3 galactosyltransferase (alpha3GT) in a complex with p-nitrophenyl-beta-galactoside (pNP-beta-Gal) | | Descriptor: | 4-nitrophenyl beta-D-galactopyranoside, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jamaluddin, H, Tumbale, P, Ferns, T.A, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2009-04-28 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Alpha-1,3 Galactosyltransferase (Alpha3Gt) in a Complex with P-Nitrophenyl-Beta-Galactoside (Pnpbetagal).
Biochem.Biophys.Res.Commun., 385, 2009
|
|
5PGU
 
 | |
7JPV
 
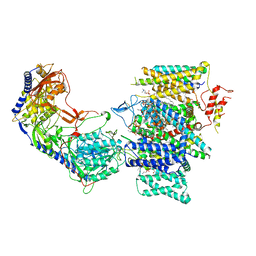 | | Rabbit Cav1.1 in the presence of 1 micromolar (S)-(-)-Bay K8644 in nanodiscs at 3.4 Angstrom resolution | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, ... | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-08-10 | | Release date: | 2020-11-18 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca v 1.1 by Dihydropyridine Compounds*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4O6V
 
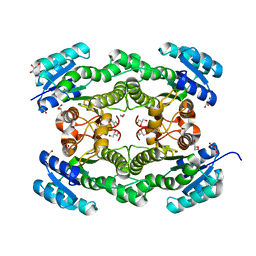 | |
3HGI
 
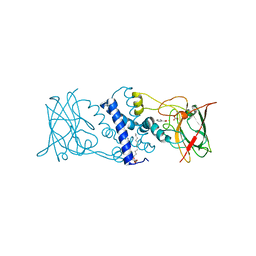 | | Crystal structure of Catechol 1,2-Dioxygenase from the gram-positive Rhodococcus opacus 1CP | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BENZOIC ACID, CARBONATE ION, ... | | Authors: | Ferraroni, M, Matera, I, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-14 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
5OOI
 
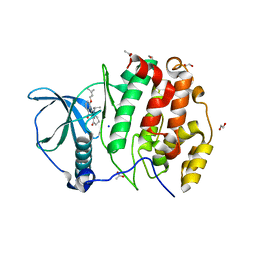 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA') IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, ACETATE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-07 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
