5YSE
 
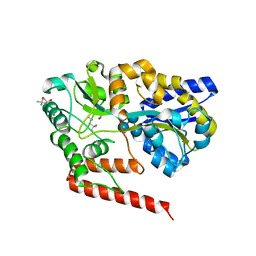 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
4IH2
 
 | | Crystal structure of kirola (Act d 11) from crystal soaked with 2-aminopurine | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
6XTU
 
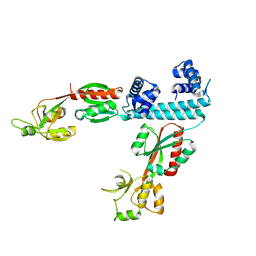 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM | | Descriptor: | Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
8VAE
 
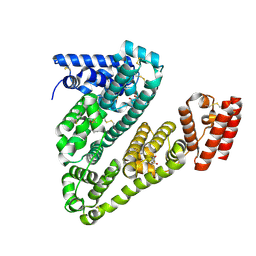 | | Cryogenic electron microscopy structure of human serum albumin in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
8VAF
 
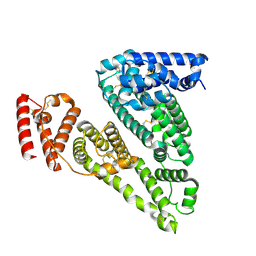 | | Cryogenic electron microscopy structure of apo human serum albumin | | Descriptor: | Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
7YUJ
 
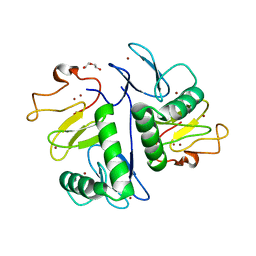 | | Crystal structure of HOIL-1L(365-510) | | Descriptor: | DI(HYDROXYETHYL)ETHER, RanBP-type and C3HC4-type zinc finger-containing protein 1, ZINC ION | | Authors: | Xiao, L, Pan, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Mechanistic insights into the enzymatic activity of E3 ligase HOIL-1L and its regulation by the linear ubiquitin chain binding.
Sci Adv, 9, 2023
|
|
8VAC
 
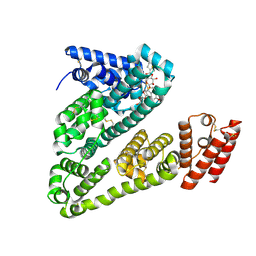 | | Cryogenic electron microscopy structure of human serum albumin in complex with teniposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-(thiophen-2-ylmethylidene)-beta-D-glucopyranoside, Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
1IL4
 
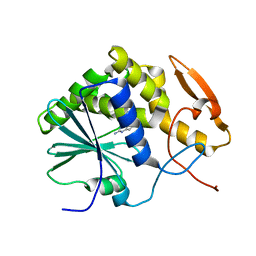 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 9-DEAZAGUANINE | | Descriptor: | 9-DEAZAGUANINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
8G2N
 
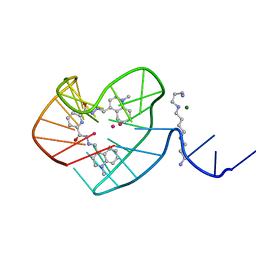 | |
8GPW
 
 | | Structure of Penicillin-binding protein 3 (PBP3) from Klebsiella pneumoniae with ligand 18G | | Descriptor: | 1-[(~{Z})-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2~{S})-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxycyclopropane-1-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Song, D.Q, Li, Y.H. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of Penicillin-binding protein 3 (PBP3) from Klebsiella pneumoniae with ligand 18G at 2.06 Angstroms resolution
To Be Published
|
|
8BQ7
 
 | |
1N8U
 
 | | Chemosensory Protein in Complex with bromo-dodecanol | | Descriptor: | BROMO-DODECANOL, chemosensory protein | | Authors: | Campanacci, V, Lartigue, A, Hallberg, B.M, Jones, T.A, Giudici-Orticoni, M.T, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N8V
 
 | | Chemosensory Protein in complex with bromo-dodecanol | | Descriptor: | BROMO-DODECANOL, chemosensory protein | | Authors: | Campanacci, V, Lartigue, A, Hallberg, B.M, Jones, T.A, Giudici-Orticoni, M.T, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2PFH
 
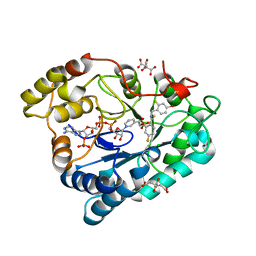 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution is less than concentration of IDD594. | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
3PV6
 
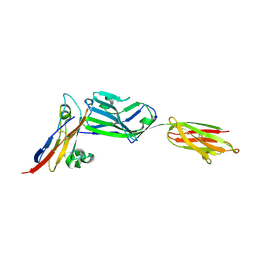 | | Crystal structure of NKp30 bound to its ligand B7-H6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig-like domain-containing protein DKFZp686O24166/DKFZp686I21167, ... | | Authors: | Li, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the activating natural killer cell receptor NKp30 bound to its ligand B7-H6 reveals basis for tumor cell recognition in humans
to be published
|
|
6WQ7
 
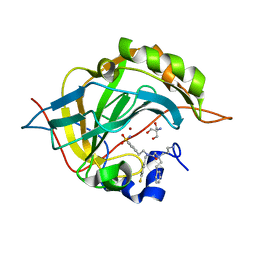 | | Carbonic Anhydrase II Complexed with 2-((3-Aminopropyl)(phenethyl)amino)-N-(4-fluorobenzyl)-N-(4-sulfamoylphenethyl)acetamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, N~2~-(3-aminopropyl)-N-[(4-fluorophenyl)methyl]-N~2~-(2-phenylethyl)-N-[2-(4-sulfamoylphenyl)ethyl]glycinamide, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
6YIM
 
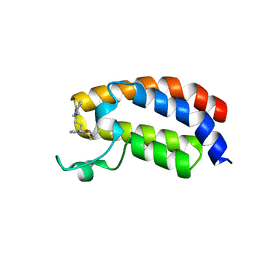 | | Crystal structure of the CREBBP bromodomain in complex with a benzo-diazepine ligand | | Descriptor: | (4~{R})-~{N}-[3-(7-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)propyl]-4-methyl-2-oxidanylidene-1,3,4,5-tetrahydro-1,5-benzodiazepine-6-carboxamide, CREBBP | | Authors: | Picaud, S, Brand, M, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Conway, S, Filippakopoulos, P. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal structure of the CREBBP bromodomain in complex with a benzo-diazepine ligand
To Be Published
|
|
3HKR
 
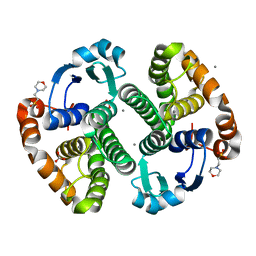 | | Crystal Structure of Glutathione Transferase Pi Y108V Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-05-25 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: structure-thermodynamic relationships and thermal stability.
Protein Sci., 18, 2009
|
|
7YNX
 
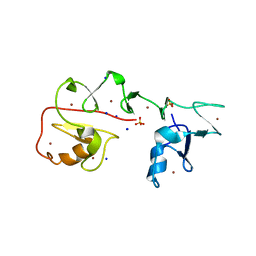 | | Crystal structure of Pirh2 bound to poly-Ala peptide | | Descriptor: | RING finger and CHY zinc finger domain-containing protein 1, SODIUM ION, SULFATE ION, ... | | Authors: | Dong, C, Yan, X, Li, Y. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of an Ala-rich C-degron by the E3 ligase Pirh2.
Nat Commun, 14, 2023
|
|
6OOR
 
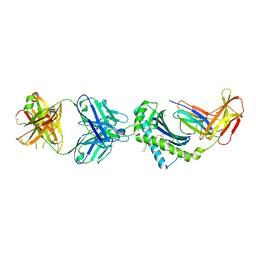 | | Structure of 1B1 bound to mouse CD1d | | Descriptor: | Antibody 1B1 Heavy chain, Antibody 1B1 Light chain, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Ying, G, Zajonc, D.M. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of NKT cell inhibition using the T-cell receptor-blocking anti-CD1d antibody 1B1.
J.Biol.Chem., 294, 2019
|
|
3GCJ
 
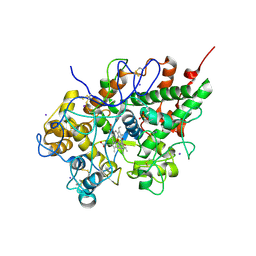 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid
To be Published
|
|
3GCK
 
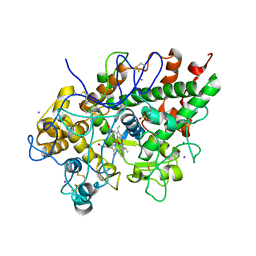 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid
To be Published
|
|
4ZYB
 
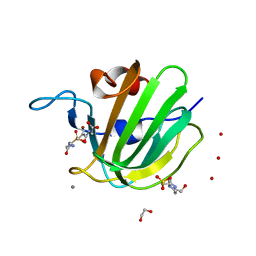 | | High resolution structure of M23 peptidase LytM with substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Grabowska, M, Jagielska, E, Czapinska, H, Bochtler, M, Sabala, I. | | Deposit date: | 2015-05-21 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution structure of an M23 peptidase with a substrate analogue.
Sci Rep, 5, 2015
|
|
3PV7
 
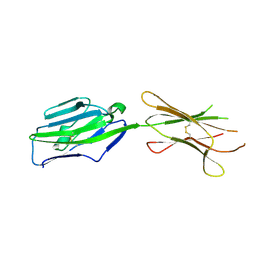 | | Crystal structure of NKp30 ligand B7-H6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig-like domain-containing protein DKFZp686O24166/DKFZp686I21167 | | Authors: | Li, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the activating natural killer cell receptor NKp30 bound to its ligand B7-H6 reveals basis for tumor cell recognition in humans
to be published
|
|
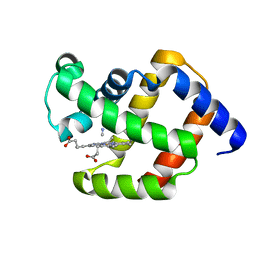
1ECN
 
 | |
