1DNK
 
 | | THE X-RAY STRUCTURE OF THE DNASE I-D(GGTATACC)2 COMPLEX AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*GP*GP*TP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3'), ... | | Authors: | Weston, S.A, Lahm, A, Suck, D. | | Deposit date: | 1992-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution.
J.Mol.Biol., 226, 1992
|
|
5FPW
 
 | |
1CUO
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ISOMER-2 AZURIN FROM METHYLOMONAS J | | Descriptor: | COPPER (II) ION, PROTEIN (AZURIN ISO-2) | | Authors: | Inoue, T, Nishio, N, Kai, Y, Suzuki, S, Kataoka, K. | | Deposit date: | 1999-08-21 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J.
J.Mol.Biol., 333, 2003
|
|
4C12
 
 | | X-ray Crystal Structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fulop, V, Roper, D.I, Ruane, K.M, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity Determinants for Lysine Incorporation in Staphylococcus Aureus Peptidoglycan as Revealed by the Structure of a Mure Enzyme Ternary Complex.
J.Biol.Chem., 288, 2013
|
|
1CHC
 
 | |
1SI2
 
 | |
1BMP
 
 | | BONE MORPHOGENETIC PROTEIN-7 | | Descriptor: | BONE MORPHOGENETIC PROTEIN-7 | | Authors: | Griffith, D.L, Scott, D.L. | | Deposit date: | 1995-12-14 | | Release date: | 1997-07-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor beta superfamily.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
4XAZ
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase variant PTE-R18, ZINC ION | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
5HDO
 
 | |
6B1R
 
 | |
7R0J
 
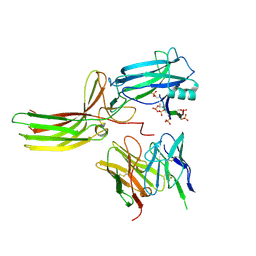 | | Structure of the V2 receptor Cter-arrestin2-ScFv30 complex | | Descriptor: | Arrestin2, ScFv30, V2R Cter | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
7R0C
 
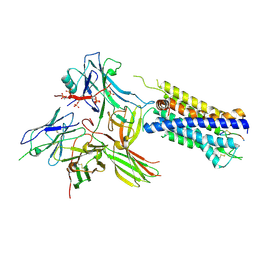 | | Structure of the AVP-V2R-arrestin2-ScFv30 complex | | Descriptor: | AVP, Arrestin2, ScFv30, ... | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
4OI0
 
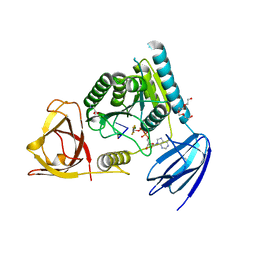 | | bound to ssRNA dinucleotide GC, ADP, AlF4-, and Mg2+(transition state, data set I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NONAETHYLENE GLYCOL, ... | | Authors: | Dikfidan, A, Loll, B, Zeymer, C, Clausen, T, Meinhart, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase clp1.
Mol.Cell, 54, 2014
|
|
4K4O
 
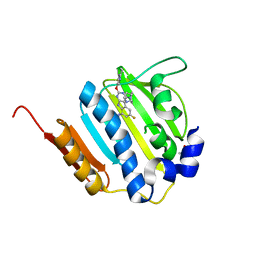 | | The DNA Gyrase B ATP binding domain of Enterococcus faecalis in complex with a small molecule inhibitor | | Descriptor: | 6-fluoro-4-[(3aR,6aR)-hexahydropyrrolo[3,4-b]pyrrol-5(1H)-yl]-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | Authors: | Bensen, D.C, Akers-Rodriguez, S, Lam, T, Tari, L.W. | | Deposit date: | 2013-04-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A new class of type IIA topoisomerase inhibitors with broad-spectrum antibacterial
activity
To be Published
|
|
2VHX
 
 | |
1HMH
 
 | |
8RHL
 
 | | Yeast 20S proteasome in complex with a linear biarylether epoxyketone (compound 15a) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Linear biarylether epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
1K94
 
 | | Crystal structure of des(1-52)grancalcin with bound calcium | | Descriptor: | CALCIUM ION, GRANCALCIN | | Authors: | Jia, J, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2001-10-26 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Ca(2+)-loaded human grancalcin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4JJB
 
 | |
1EFC
 
 | | INTACT ELONGATION FACTOR FROM E.COLI | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (ELONGATION FACTOR) | | Authors: | Song, H, Parsons, M.R, Rowsell, S, Leonard, G, Phillips, S.E.V. | | Deposit date: | 1998-11-24 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution.
J.Mol.Biol., 285, 1999
|
|
1EGO
 
 | | NMR STRUCTURE OF OXIDIZED ESCHERICHIA COLI GLUTAREDOXIN: COMPARISON WITH REDUCED E. COLI GLUTAREDOXIN AND FUNCTIONALLY RELATED PROTEINS | | Descriptor: | GLUTAREDOXIN | | Authors: | Xia, T.-H, Bushweller, J.H, Sodano, P, Billeter, M, Bjornberg, O, Holmgren, A, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and functionally related proteins.
Protein Sci., 1, 1992
|
|
5HVS
 
 | | Crystal Structure of Macrophage Migration Inhibitory Factor (MIF) with a Biaryltriazole Inhibitor (3i-305) | | Descriptor: | 3-({2-[1-(3-fluoro-4-hydroxyphenyl)-1H-1,2,3-triazol-4-yl]quinolin-5-yl}oxy)benzoic acid, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2016-01-28 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Fluorescence Polarization Assay for Binding to Macrophage Migration Inhibitory Factor and Crystal Structures for Complexes of Two Potent Inhibitors.
J.Am.Chem.Soc., 138, 2016
|
|
6B1S
 
 | |
1EGR
 
 | | SEQUENCE-SPECIFIC 1H N.M.R. ASSIGNMENTS AND DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF REDUCED ESCHERICHIA COLI GLUTAREDOXIN | | Descriptor: | GLUTAREDOXIN | | Authors: | Sodano, P, Xia, T.-H, Bushweller, J.H, Bjornberg, O, Holmgren, A, Billeter, M, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific 1H n.m.r. assignments and determination of the three-dimensional structure of reduced Escherichia coli glutaredoxin.
J.Mol.Biol., 221, 1991
|
|
1EPU
 
 | | X-RAY crystal structure of neuronal SEC1 from squid | | Descriptor: | S-SEC1 | | Authors: | Bracher, A, Perrakis, A, Dresbach, T, Betz, H, Weissenhorn, W. | | Deposit date: | 2000-03-29 | | Release date: | 2000-08-09 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray crystal structure of neuronal Sec1 from squid sheds new light on the role of this protein in exocytosis.
Structure Fold.Des., 8, 2000
|
|
