8VQU
 
 | | Crystal Structure of Dehaloperoxidase B in complex with substrate 1,4-cyclohexadiene | | Descriptor: | Dehaloperoxidase B, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | de Seerano, V.S, Yun, D, Ghiladi, R.A. | | Deposit date: | 2024-01-19 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal Structure of Dehaloperoxidase B in complex with substrate 1,4-cyclohexadiene
To Be Published
|
|
4XUN
 
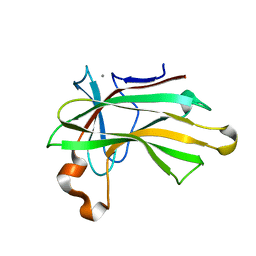 | |
6P35
 
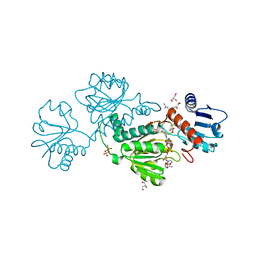 | |
6P6R
 
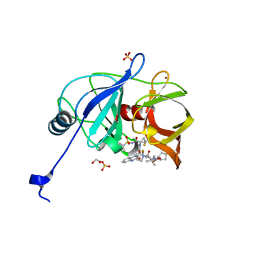 | |
7QE3
 
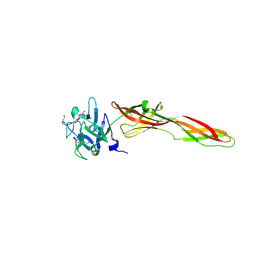 | | Se-M variant of B-trefoil lectin from Salpingoeca rosetta in complex with GalNAc | | Descriptor: | (2S)-hexane-1,2,6-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Notova, S, Varrot, A. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The choanoflagellate pore-forming lectin SaroL-1 punches holes in cancer cells by targeting the tumor-related glycosphingolipid Gb3.
Commun Biol, 5, 2022
|
|
4Y8X
 
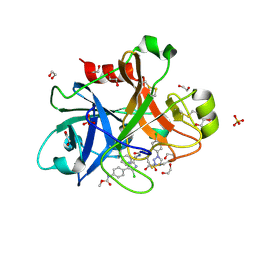 | |
7CDH
 
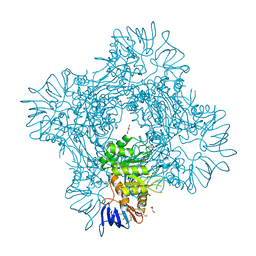 | | Crystal structure of Betaaspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum-AW-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Isoaspartyl dipeptidase, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
6QZB
 
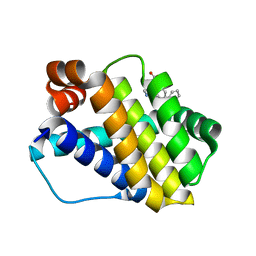 | | Structure of Mcl-1 in complex with compound 8d | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(2-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6P26
 
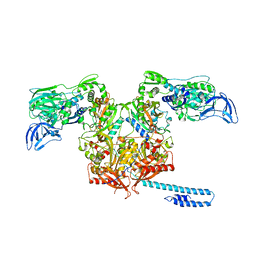 | | Escherichia coli tRNA synthetase in complex with compound 1 | | Descriptor: | N-benzyl-2-(cyclohex-1-en-1-yl)ethan-1-amine, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Kahne, D, Baidin, V, Owens, T.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Simple Secondary Amines Inhibit Growth of Gram-Negative Bacteria through Highly Selective Binding to Phenylalanyl-tRNA Synthetase.
J.Am.Chem.Soc., 143, 2021
|
|
8FR5
 
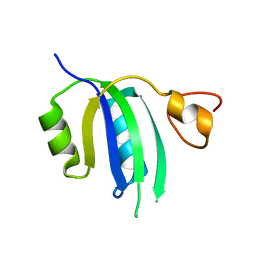 | | Crystal structure of the Human Smacovirus 1 Rep domain | | Descriptor: | MANGANESE (II) ION, Rep, SODIUM ION | | Authors: | Limon, L.K, Shi, K, Dao, A, Rugloski, J, Tompkins, K.J, Aihara, H, Gordon, W.R, Evans IIII, R.L. | | Deposit date: | 2023-01-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The crystal structure of the human smacovirus 1 Rep domain.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7PTH
 
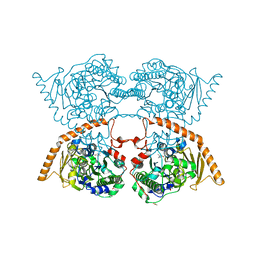 | |
8RMW
 
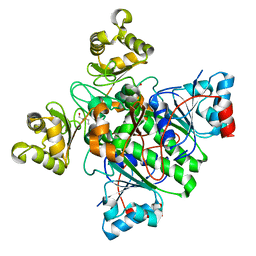 | | Alpha-Methylacyl-CoA racemase from Mycobacterium tuberculosis. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-methylacyl-CoA racemase, DI(HYDROXYETHYL)ETHER | | Authors: | Mojanaga, O.O, Acharya, K.R, Lloyd, M.D. | | Deposit date: | 2024-01-09 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | alpha-Methylacyl-CoA Racemase from Mycobacterium tuberculosis -Detailed Kinetic and Structural Characterization of the Active Site.
Biomolecules, 14, 2024
|
|
6P3P
 
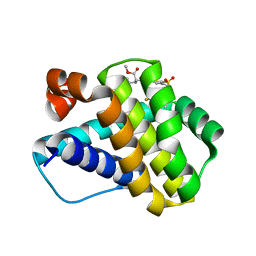 | | Crystal structure of Mcl-1 in complex with compound 65 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, methyl N-(5-{[2-chloro-5-(trifluoromethyl)phenyl]sulfamoyl}-4-methylthiophene-2-carbonyl)-D-phenylalaninate | | Authors: | Toms, A.V, Follows, B. | | Deposit date: | 2019-05-24 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of novel biaryl sulfonamide based Mcl-1 inhibitors.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4YDJ
 
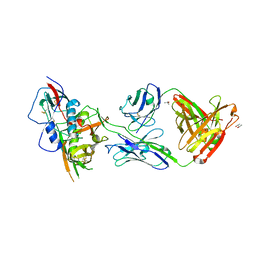 | | Crystal structure of broadly and potently neutralizing antibody 44-VRC13.01 in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
9SMQ
 
 | | Crystal structure of LRH-1/TIF-2 peptide in complex with CP4 | | Descriptor: | 3-[5-phenyl-1-[3-(trifluoromethyl)phenyl]pyrazol-3-yl]propanoic acid, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2, ... | | Authors: | Kim, Y, Ildefeld, N, Heering, J, Proschak, E, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-09-08 | | Release date: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of LRH-1/TIF-2 peptide in complex with CP4
To Be Published
|
|
7MC6
 
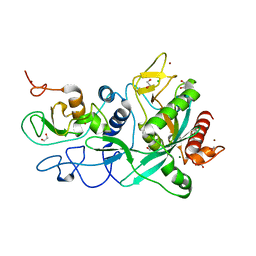 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex containing Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MC5
 
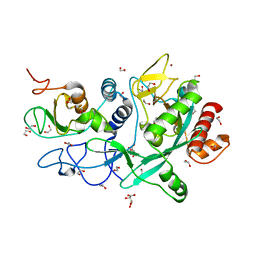 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8X47
 
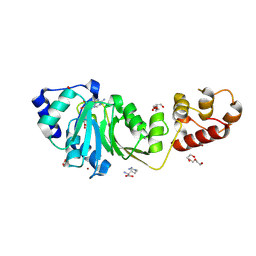 | | Crystal structure of DIMT1 in complex with S-adenosyl-L-homocysteine (SAH) from Pyrococcus horikoshii | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saha, S, Dutta, A, Kanaujia, S.P. | | Deposit date: | 2023-11-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional characterization of archaeal DIMT1 unveils distinct protein dynamics essential for efficient catalysis.
Structure, 32, 2024
|
|
1AFW
 
 | |
7NBP
 
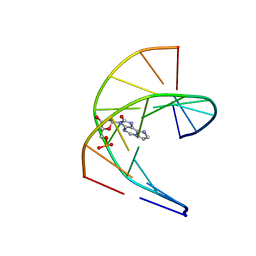 | | Solution structure of DNA duplex containing a 7,8-dihydro-8-oxo-1,N6-ethenoadenine base modification that induces exclusively A->T transversions in Escherichia coli | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*(EDA)P*CP*AP*TP*G)-3') | | Authors: | Aralov, A.V, Gubina, N, Cabrero, C, Tsvetkov, V.B, Turaev, A.V, Fedeles, B.I, Croy, R.G, Isaakova, E.A, Melnik, D, Dukova, S, Khrulev, A.A, Varizhuk, A.M, Gonzalez, C, Zatsepin, T.S, Essigmann, J.M. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 7,8-Dihydro-8-oxo-1,N6-ethenoadenine: an exclusively Hoogsteen-paired thymine mimic in DNA that induces A→T transversions in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
6EDV
 
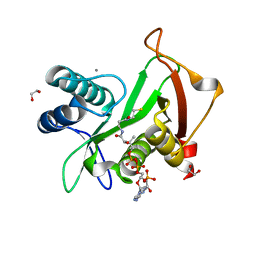 | | Structure of a GNAT superfamily acetyltransferase PA3944 in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, CALCIUM ION, ... | | Authors: | Majorek, K.A, Satchell, K.J.F, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-12 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro.
Biochemistry, 57, 2018
|
|
8JU0
 
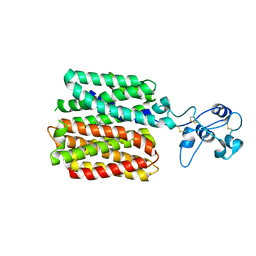 | | hOCT1 in complex with spironolactone in inward facing occluded conformation | | Descriptor: | SPIRONOLACTONE, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-27 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTT
 
 | | hOCT1 in complex with metformin in outward occluded conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTS
 
 | | hOCT1 in complex with metformin in outward open conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTV
 
 | | hOCT1 in complex with metformin in inward occluded conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
