4LVZ
 
 | |
4MHH
 
 | |
4LRD
 
 | | Phosphopentomutase 4H11 variant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LVY
 
 | | Structure of the THF riboswitch bound to pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
4LRC
 
 | | Phosphopentomutase V158L variant | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LUD
 
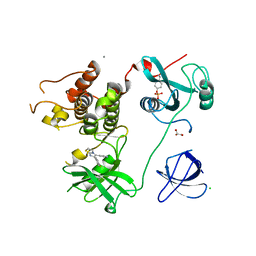 | | Crystal Structure of HCK in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LR7
 
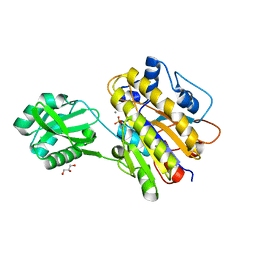 | | Phosphopentomutase S154A variant | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LVV
 
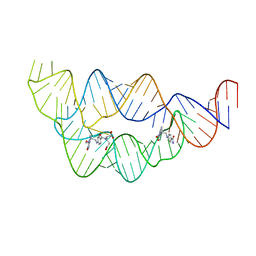 | | Structure of the THF riboswitch | | Descriptor: | N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
2VEA
 
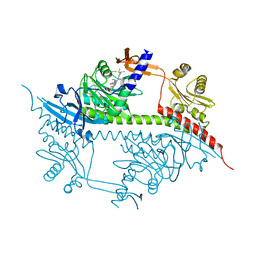 | |
4LRB
 
 | | Phosphopentomutase S154G variant soaked with 2,3-dideoxyribose 5-phosphate | | Descriptor: | 2,3-dideoxy-5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, GLYCEROL, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LR8
 
 | | Phosphopentomutase S154A variant soaked with ribose 5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
7UFG
 
 | | Cryo-EM structure of PAPP-A in complex with IGFBP5 | | Descriptor: | Insulin-like growth factor-binding protein 5, Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
7UKK
 
 | |
7UJU
 
 | | Room-temperature X-ray structure of monomeric SARS-CoV-2 main protease catalytic domain (MPro1-196) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-03-31 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Autoprocessing and oxyanion loop reorganization upon GC373 and nirmatrelvir binding of monomeric SARS-CoV-2 main protease catalytic domain.
Commun Biol, 5, 2022
|
|
7UJ9
 
 | |
7UJG
 
 | |
3P19
 
 | | Improved NADPH-dependent Blue Fluorescent Protein | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative blue fluorescent protein | | Authors: | Kao, T.H, Chen, Y, Pai, C.H, Wang, A.H.J. | | Deposit date: | 2010-09-30 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a NADPH-dependent blue fluorescent protein revealed the unique role of Gly176 on the fluorescence enhancement.
J.Struct.Biol., 174, 2011
|
|
7UJ4
 
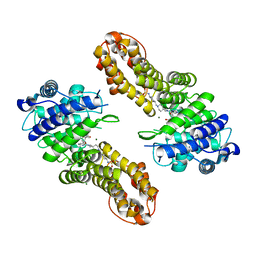 | | Inhibition of Human Menin by SNDX-5613 | | Descriptor: | 2-({4-[7-({(1r,4r)-4-[(ethanesulfonyl)amino]cyclohexyl}methyl)-2,7-diazaspiro[3.5]nonan-2-yl]pyrimidin-5-yl}oxy)-N-ethyl-5-fluoro-N-(propan-2-yl)benzamide, Isoform 2 of Menin, MAGNESIUM ION | | Authors: | McKeever, B.M, Kulkarni, S, McGeehan, G.M. | | Deposit date: | 2022-03-30 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | MEN1 mutations mediate clinical resistance to menin inhibition.
Nature, 615, 2023
|
|
3OPZ
 
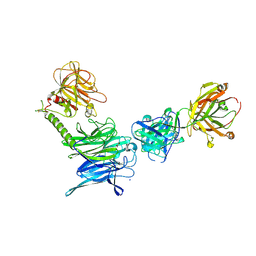 | | Crystal structure of trans-sialidase in complex with the Fab fragment of a neutralizing monoclonal IgG antibody | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, Trans-sialidase, ... | | Authors: | Larrieux, N, Muia, R, Campetella, O, Buschiazzo, A. | | Deposit date: | 2010-09-02 | | Release date: | 2011-11-09 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Trypanosoma cruzi trans-sialidase in complex with a neutralizing antibody: structure/function studies towards the rational design of inhibitors.
Plos Pathog., 8, 2012
|
|
4LFS
 
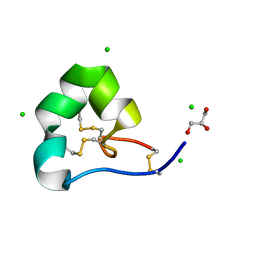 | | High resolution x-ray structure of racemic ShK toxin | | Descriptor: | CHLORIDE ION, GLYCEROL, Potassium channel toxin ShK | | Authors: | Dang, B, Kubota, T, Mandal, K, Bezanilla, F, Kent, S.B.H. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Native Chemical Ligation at Asx-Cys, Glx-Cys: Chemical Synthesis and High-Resolution X-ray Structure of ShK Toxin by Racemic Protein Crystallography.
J.Am.Chem.Soc., 135, 2013
|
|
4DVX
 
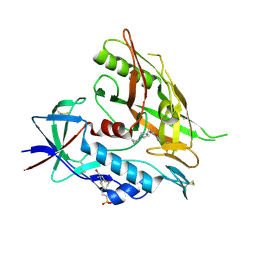 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with MAE-II-188 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-(4-chloro-3-fluorophenyl)-N'-{[(3R)-1-cyclopropylpyrrolidin-3-yl]methyl}ethanediamide, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2012-02-23 | | Release date: | 2013-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of HIV-1 gp120 Envelope Glycoprotein in Complex with NBD Analogues That Target the CD4-Binding Site.
Plos One, 9, 2014
|
|
3OLA
 
 | | Poliovirus polymerase elongation complex with 2'-deoxy-CTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA/RNA (5'-R(*GP*CP*CP*CP*GP*GP*AP*CP*GP*AP*GP*AP*GP*A)-D(P*C)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2010-08-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4LR9
 
 | | Phosphopentomutase S154A variant soaked with 2,3-dideoxyribose 5-phosphate | | Descriptor: | 2,3-dideoxy-5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Birmingham, W.A, Starbird, C.A, Panosian, T.D, Nannemann, D.P, Iverson, T.M, Bachmann, B.O. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bioretrosynthetic construction of a didanosine biosynthetic pathway.
Nat.Chem.Biol., 10, 2014
|
|
4LVW
 
 | |
4LUE
 
 | | Crystal Structure of HCK in complex with 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (resulting from displacement of SKF86002) | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
